A novel Bcl-2-like inhibitor of apoptosis is encoded by the parapoxvirus ORF virus
- PMID: 17475653
- PMCID: PMC1933275
- DOI: 10.1128/JVI.00404-07
A novel Bcl-2-like inhibitor of apoptosis is encoded by the parapoxvirus ORF virus
Abstract
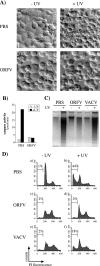
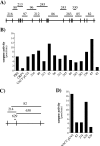
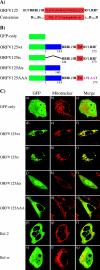
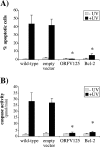
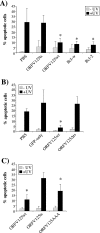
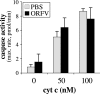
Apoptotic cell death forms part of the host defense against virus infection. We tested orf virus, a member of the poxvirus family, for the ability to inhibit apoptosis and found that orf virus-infected cells were fully resistant to UV-induced changes in cell morphology, caspase activation, and DNA fragmentation. By using a library of vaccinia virus-orf virus recombinants, we identified an orf virus gene (ORFV125) whose presence was linked with the inhibition of apoptosis. The 173-amino-acid predicted protein had no clear homologs in public databases other than those encoded by other parapoxviruses. However, ORFV125 possessed a distinctive C-terminal domain which was necessary and sufficient to direct the protein to the mitochondria. We determined that ORFV125 alone could fully inhibit UV-induced DNA fragmentation, caspase activation, and cytochrome c release and that its mitochondrial localization was required for its antiapoptotic function. In contrast, ORFV125 did not prevent UV-induced activation of c-Jun NH2-terminal kinase, an event occurring upstream of the mitochondria. These features are comparable to the antiapoptotic properties of the mitochondrial regulator Bcl-2. Furthermore, bioinformatic analyses revealed sequence and secondary-structure similarities to Bcl-2 family members, including characteristic residues of all four Bcl-2 homology domains. Consistent with this, the viral protein inhibited the UV-induced activation of the proapoptotic Bcl-2 family members Bax and Bak. ORFV125 is the first parapoxvirus apoptosis inhibitor to be identified, and we propose that it is a new antiapoptotic member of the Bcl-2 family.
Figures
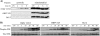


Similar articles
-
Ionizing radiation utilizes c-Jun N-terminal kinase for amplification of mitochondrial apoptotic cell death in human cervical cancer cells.FEBS J. 2008 May;275(9):2096-108. doi: 10.1111/j.1742-4658.2008.06363.x. Epub 2008 Mar 28. FEBS J. 2008. PMID: 18373696
-
Cytomegaloviruses inhibit Bak- and Bax-mediated apoptosis with two separate viral proteins.Cell Death Differ. 2010 Apr;17(4):655-65. doi: 10.1038/cdd.2009.147. Epub 2009 Oct 9. Cell Death Differ. 2010. PMID: 19816509
-
Fowlpox virus encodes a Bcl-2 homologue that protects cells from apoptotic death through interaction with the proapoptotic protein Bak.J Virol. 2007 Oct;81(20):11032-45. doi: 10.1128/JVI.00734-07. Epub 2007 Aug 8. J Virol. 2007. PMID: 17686864 Free PMC article.
-
Bcl-2 family proteins: the sentinels of the mitochondrial apoptosis pathway.IUBMB Life. 2008 Jun;60(6):390-7. doi: 10.1002/iub.51. IUBMB Life. 2008. PMID: 18425793 Review.
-
Mechanisms of apoptosis regulation by viral oncogenes in infection and tumorigenesis.Cell Death Differ. 2006 Aug;13(8):1371-7. doi: 10.1038/sj.cdd.4401941. Epub 2006 May 5. Cell Death Differ. 2006. PMID: 16676007 Review.
Cited by
-
Structural Investigation of Orf Virus Bcl-2 Homolog ORFV125 Interactions with BH3-Motifs from BH3-Only Proteins Puma and Hrk.Viruses. 2021 Jul 15;13(7):1374. doi: 10.3390/v13071374. Viruses. 2021. PMID: 34372579 Free PMC article.
-
How do viruses control mitochondria-mediated apoptosis?Virus Res. 2015 Nov 2;209:45-55. doi: 10.1016/j.virusres.2015.02.026. Epub 2015 Mar 1. Virus Res. 2015. PMID: 25736565 Free PMC article. Review.
-
The vaccinia virus-encoded Bcl-2 homologues do not act as direct Bax inhibitors.J Virol. 2012 Jan;86(1):203-13. doi: 10.1128/JVI.05817-11. Epub 2011 Oct 19. J Virol. 2012. PMID: 22013032 Free PMC article.
-
Functional analysis of the short isoform of orf virus protein OV20.0.J Virol. 2015 May;89(9):4966-79. doi: 10.1128/JVI.03714-14. Epub 2015 Feb 18. J Virol. 2015. PMID: 25694596 Free PMC article.
-
A nuclear inhibitor of NF-kappaB encoded by a poxvirus.J Virol. 2011 Jan;85(1):264-75. doi: 10.1128/JVI.01149-10. Epub 2010 Oct 27. J Virol. 2011. PMID: 20980501 Free PMC article.
References
-
- Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. - PubMed
-
- Breckenridge, D. G., and D. Xue. 2004. Regulation of mitochondrial membrane permeabilization by BCL-2 family proteins and caspases. Curr. Opin. Cell Biol. 16:647-652. - PubMed
-
- Brick, D. J., R. D. Burke, A. A. Minkley, and C. Upton. 2000. Ectromelia virus virulence factor p28 acts upstream of caspase-3 in response to UV light-induced apoptosis. J. Gen. Virol. 81:1087-1097. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

