Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China
- PMID: 17459938
- PMCID: PMC1933311
- DOI: 10.1128/JVI.00299-07
Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China
Abstract
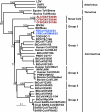
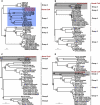
Since an outbreak of severe acute respiratory syndrome (SARS) was averted in 2004, many novel coronaviruses have been recognized from different species, including humans. Bats have provided the most diverse assemblages of coronaviruses, suggesting that they may be the natural reservoir. Continued virological surveillance has proven to be the best way to avert this infectious disease at the source. Here we provide the first description of a previously unidentified coronavirus lineage detected from wild Asian leopard cats (Prionailurus bengalensis) and Chinese ferret badgers (Melogale moschata) during virological surveillance in southern China. Partial genome analysis revealed a typical coronavirus genome but with a unique putative accessory gene organization. Phylogenetic analyses revealed that the envelope, membrane, and nucleoprotein structural proteins and the two conserved replicase domains, putative RNA-dependent RNA polymerase and RNA helicase, of these novel coronaviruses were most closely related to those of group 3 coronaviruses identified from birds, while the spike protein gene was most closely related to that of group 1 coronaviruses from mammals. However, these viruses always fell into an outgroup phylogenetic relationship with respect to other coronaviruses and had low amino acid similarity to all known coronavirus groups, indicating that they diverged early in the evolutionary history of coronaviruses. These results suggest that these viruses may represent a previously unrecognized evolutionary pathway, or possibly an unidentified coronavirus group. This study demonstrates the importance of systematic virological surveillance in market animals for understanding the evolution and emergence of viruses with infectious potential.
Figures


Similar articles
-
Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus.J Virol. 2009 Jan;83(2):908-17. doi: 10.1128/JVI.01977-08. Epub 2008 Oct 29. J Virol. 2009. PMID: 18971277 Free PMC article.
-
SARS/avian coronaviruses.Dev Biol (Basel). 2006;126:161-9; discussion 326-7. Dev Biol (Basel). 2006. PMID: 17058491
-
Prevalence and genetic diversity of coronaviruses in bats from China.J Virol. 2006 Aug;80(15):7481-90. doi: 10.1128/JVI.00697-06. J Virol. 2006. PMID: 16840328 Free PMC article.
-
Coronavirus diversity, phylogeny and interspecies jumping.Exp Biol Med (Maywood). 2009 Oct;234(10):1117-27. doi: 10.3181/0903-MR-94. Epub 2009 Jun 22. Exp Biol Med (Maywood). 2009. PMID: 19546349 Review.
-
Coronaviruses in poultry and other birds.Avian Pathol. 2005 Dec;34(6):439-48. doi: 10.1080/03079450500367682. Avian Pathol. 2005. PMID: 16537157 Review.
Cited by
-
NLRP1 restricts porcine deltacoronavirus infection via IL-11 inhibiting the phosphorylation of the ERK signaling pathway.J Virol. 2024 Mar 19;98(3):e0198223. doi: 10.1128/jvi.01982-23. Epub 2024 Feb 27. J Virol. 2024. PMID: 38411106 Free PMC article.
-
Research Advances on Swine Acute Diarrhea Syndrome Coronavirus.Animals (Basel). 2024 Jan 30;14(3):448. doi: 10.3390/ani14030448. Animals (Basel). 2024. PMID: 38338091 Free PMC article. Review.
-
Binding affinity between coronavirus spike protein and human ACE2 receptor.Comput Struct Biotechnol J. 2024 Jan 17;23:759-770. doi: 10.1016/j.csbj.2024.01.009. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38304547 Free PMC article. Review.
-
Prevention and Control of Swine Enteric Coronaviruses in China: A Review of Vaccine Development and Application.Vaccines (Basel). 2023 Dec 21;12(1):11. doi: 10.3390/vaccines12010011. Vaccines (Basel). 2023. PMID: 38276670 Free PMC article. Review.
-
Lack of detection of SARS-CoV-2 in British wildlife 2020-21 and first description of a stoat (Mustela erminea) Minacovirus.J Gen Virol. 2023 Dec;104(12):001917. doi: 10.1099/jgv.0.001917. J Gen Virol. 2023. PMID: 38059490 Free PMC article.
References
-
- Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. M. Peiris, and L. L. M. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. - PubMed
-
- Guan, Y., N. S. Zhong, H. Chen, G. J. D. Smith, and B. J. Zheng. 2006. An averted SARS outbreak, p. 93-100. In J. C. K. Chan and V. C. W. Taam Wong (ed.) Challenges of severe acute respiratory syndrome. Elsevier, New York, NY.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

