Analysis of the genome sequence of an alpaca coronavirus
- PMID: 17459444
- PMCID: PMC7185508
- DOI: 10.1016/j.virol.2007.03.035
Analysis of the genome sequence of an alpaca coronavirus
Abstract
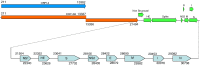
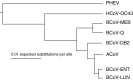
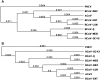
Coronaviral infection of New World camelids was first identified in 1998 in llamas and alpacas with severe diarrhea. In order to understand this infection, one of the coronavirus isolates was sequenced and analyzed. It has a genome of 31,076 nt including the poly A tail at the 3' end. This virus designated as ACoV-00-1381 (ACoV) encodes all 10 open reading frames (ORFs) characteristic of Group 2 bovine coronavirus (BCoV). Phylogenetic analysis showed that the ACoV genome is clustered closely (>99.5% identity) with two BCoV strains, ENT and LUN, and was also closely related to other BCoV strains (Mebus, Quebec, DB2), a human corona virus (strain 043) (>96%), and porcine hemagglutinating encephalomyelitis virus (>93% identity). A total of 145 point mutations and one nucleotide deletion were found relative to the BCoV ENT. Most of the ORFs were highly conserved; however, the predicted spike protein (S) has 9 and 12 amino acid differences from BCoV LUN and ENT, respectively, and shows a higher relative number of changes than the other proteins. Phylogenetic analysis suggests that ACoV shares the same ancestor as BCoV ENT and LUN.
Figures
Similar articles
-
Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe.J Virol. 2007 May;81(10):4981-90. doi: 10.1128/JVI.02361-06. Epub 2007 Mar 7. J Virol. 2007. PMID: 17344285 Free PMC article.
-
Molecular analysis of Brazilian strains of bovine coronavirus (BCoV) reveals a deletion within the hypervariable region of the S1 subunit of the spike glycoprotein also found in human coronavirus OC43.Arch Virol. 2006 Sep;151(9):1735-48. doi: 10.1007/s00705-006-0752-9. Epub 2006 Apr 3. Arch Virol. 2006. PMID: 16583154 Free PMC article.
-
Molecular analysis of the bovine coronavirus S1 gene by direct sequencing of diarrheic fecal specimens.Braz J Med Biol Res. 2008 Apr;41(4):277-82. doi: 10.1590/s0100-879x2008000400004. Braz J Med Biol Res. 2008. PMID: 18392449
-
Genomic Characterization and Phylogenetic Classification of Bovine Coronaviruses Through Whole Genome Sequence Analysis.Viruses. 2020 Feb 6;12(2):183. doi: 10.3390/v12020183. Viruses. 2020. PMID: 32041103 Free PMC article.
-
Advances in Bovine Coronavirus Epidemiology.Viruses. 2022 May 21;14(5):1109. doi: 10.3390/v14051109. Viruses. 2022. PMID: 35632850 Free PMC article. Review.
Cited by
-
Detection of Alpha- and Betacoronaviruses in Small Mammals in Western Yunnan Province, China.Viruses. 2023 Sep 20;15(9):1965. doi: 10.3390/v15091965. Viruses. 2023. PMID: 37766371 Free PMC article.
-
Co-Circulation of Multiple Coronavirus Genera and Subgenera during an Epizootic of Lethal Respiratory Disease in Newborn Alpacas (Vicugna pacos) in Peru: First Report of Bat-like Coronaviruses in Alpacas.Animals (Basel). 2023 Sep 21;13(18):2983. doi: 10.3390/ani13182983. Animals (Basel). 2023. PMID: 37760383 Free PMC article.
-
Using machine learning to detect coronaviruses potentially infectious to humans.Sci Rep. 2023 Jun 8;13(1):9319. doi: 10.1038/s41598-023-35861-7. Sci Rep. 2023. PMID: 37291260 Free PMC article.
-
Variations in Cell Surface ACE2 Levels Alter Direct Binding of SARS-CoV-2 Spike Protein and Viral Infectivity: Implications for Measuring Spike Protein Interactions with Animal ACE2 Orthologs.J Virol. 2022 Sep 14;96(17):e0025622. doi: 10.1128/jvi.00256-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000847 Free PMC article.
-
Bovine Coronavirus Infects the Respiratory Tract of Cattle Challenged Intranasally.Front Vet Sci. 2022 Apr 29;9:878240. doi: 10.3389/fvets.2022.878240. eCollection 2022. Front Vet Sci. 2022. PMID: 35573402 Free PMC article.
References
-
- Cavanagh D., Brian D.A., Brinton M.A., Enjuanes L., Holmes K.V., Horzinek M.C., Lai M.M., Laude H., Plagemann P.G., Siddell S.G. The Coronaviridae now comprises two genera, coronavirus and torovirus: report of the Coronaviridae Study Group. Adv. Exp. Med. Biol. 1993;342:255–257. - PubMed
-
- Cebra C.K., Mattson D.E., Baker R.J., Sonn R.J., Dearing P.L. Potential pathogens in feces from unweaned llamas and alpacas with diarrhea. J. Am. Vet. Med. Assoc. 2003;223(12):1806–1808. - PubMed
-
- Che X.Y., Qiu L.W., Pan Y.X., Wen K., Hao W., Zhang L.Y., Wang Y.D., Liao Z.Y., Hua X., Cheng V.C., Yuen K.Y. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J. Clin. Microbiol. 2004;42(6):2629–2635. - PMC - PubMed
-
- Chouljenko V.N., Foster T.P., Lin X., Storz J., Kousoulas K.G. Elucidation of the genomic nucleotide sequence of bovine coronavirus and analysis of cryptic leader mRNA fusion sites. Adv. Exp. Med. Biol. 2001;494:49–55. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

