Potential interface between ribosomal protein production and pre-rRNA processing
- PMID: 17452446
- PMCID: PMC1951472
- DOI: 10.1128/MCB.02062-06
Potential interface between ribosomal protein production and pre-rRNA processing
Abstract
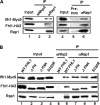
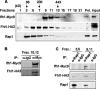
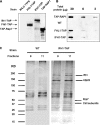
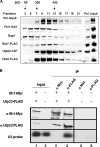
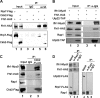
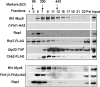
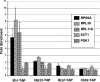
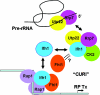
It has become clear that in Saccharomyces cerevisiae the transcription of ribosomal protein genes, which makes up a major proportion of the total transcription by RNA polymerase II, is controlled by the interaction of three transcription factors, Rap1, Fhl1, and Ifh1. Of these, only Rap1 binds directly to DNA and only Ifh1 is absent when transcription is repressed. We have examined further the nature of this interaction and find that Ifh1 is actually associated with at least two complexes. In addition to its association with Rap1 and Fhl1, Ifh1 forms a complex (CURI) with casein kinase 2 (CK2), Utp22, and Rrp7. Fhl1 is loosely associated with the CURI complex; its absence partially destabilizes the complex. The CK2 within the complex phosphorylates Ifh1 in vitro but no other members of the complex. Two major components of this complex, Utp22 and Rrp7, are essential participants in the processing of pre-rRNA. Depletion of either protein, but not of other proteins in the early processing steps, brings about a substantial increase in ribosomal protein mRNA. We propose a model in which the CURI complex is a key mediator between the two parallel pathways necessary for ribosome synthesis: the transcription and processing of pre-rRNA and the transcription of ribosomal protein genes.
Figures


Similar articles
-
A Molecular Titration System Coordinates Ribosomal Protein Gene Transcription with Ribosomal RNA Synthesis.Mol Cell. 2016 Nov 17;64(4):720-733. doi: 10.1016/j.molcel.2016.10.003. Epub 2016 Nov 3. Mol Cell. 2016. PMID: 27818142
-
Role of CK2-dependent phosphorylation of Ifh1 and Crf1 in transcriptional regulation of ribosomal protein genes in Saccharomyces cerevisiae.Biochim Biophys Acta. 2016 Aug;1859(8):1004-13. doi: 10.1016/j.bbagrm.2016.06.003. Epub 2016 Jun 15. Biochim Biophys Acta. 2016. PMID: 27321754
-
Fine-structure analysis of ribosomal protein gene transcription.Mol Cell Biol. 2006 Jul;26(13):4853-62. doi: 10.1128/MCB.02367-05. Mol Cell Biol. 2006. PMID: 16782874 Free PMC article.
-
Feedback regulation of ribosome assembly.Curr Genet. 2018 Apr;64(2):393-404. doi: 10.1007/s00294-017-0764-x. Epub 2017 Oct 11. Curr Genet. 2018. PMID: 29022131 Review.
-
Transcriptional control of ribosome biogenesis in yeast: links to growth and stress signals.Biochem Soc Trans. 2021 Aug 27;49(4):1589-1599. doi: 10.1042/BST20201136. Biochem Soc Trans. 2021. PMID: 34240738 Free PMC article. Review.
Cited by
-
Engineering the supply chain for protein production/secretion in yeasts and mammalian cells.J Ind Microbiol Biotechnol. 2015 Mar;42(3):453-64. doi: 10.1007/s10295-014-1569-2. Epub 2015 Jan 6. J Ind Microbiol Biotechnol. 2015. PMID: 25561318 Review.
-
Highly redundant function of multiple AT-rich sequences as core promoter elements in the TATA-less RPS5 promoter of Saccharomyces cerevisiae.Nucleic Acids Res. 2011 Jan;39(1):59-75. doi: 10.1093/nar/gkq741. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805245 Free PMC article.
-
Arabidopsis RIBOSOMAL RNA PROCESSING7 Is Required for 18S rRNA Maturation.Plant Cell. 2018 Nov;30(11):2855-2872. doi: 10.1105/tpc.18.00245. Epub 2018 Oct 25. Plant Cell. 2018. PMID: 30361235 Free PMC article.
-
Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs.Mol Cell Biol. 2008 May;28(10):3089-100. doi: 10.1128/MCB.01574-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332121 Free PMC article.
-
SIR telomere silencing depends on nuclear envelope lipids and modulates sensitivity to a lysolipid.J Cell Biol. 2023 Jul 3;222(7):e202206061. doi: 10.1083/jcb.202206061. Epub 2023 Apr 12. J Cell Biol. 2023. PMID: 37042812 Free PMC article.
References
-
- Barz, T., K. Ackermann, G. Dubois, R. Eils, and W. Pyerin. 2003. Genome-wide expression screens indicate a global role for protein kinase CK2 in chromatin remodeling. J. Cell Sci. 116:1563-1577. - PubMed
-
- DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. - PubMed
-
- Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
