SARS coronavirus and innate immunity
- PMID: 17451827
- PMCID: PMC2292640
- DOI: 10.1016/j.virusres.2007.03.015
SARS coronavirus and innate immunity
Abstract
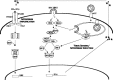
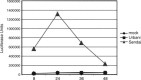
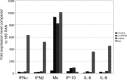
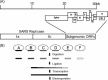
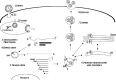
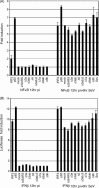
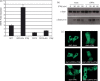
The emergence of the highly pathogenic SARS coronavirus (SARS-CoV) has reignited interest in coronavirus biology and pathogenesis. An emerging theme in coronavirus pathogenesis is that the interaction between specific viral genes and the host immune system, specifically the innate immune system, functions as a key determinant in regulating virulence and disease outcomes. Using SARS-CoV as a model, we will review the current knowledge of the interplay between coronavirus infection and the host innate immune system in vivo, and then discuss the mechanisms by which specific gene products antagonize the host innate immune response in cell culture models. Our data suggests that the SARS-CoV uses specific strategies to evade and antagonize the sensing and signaling arms of the interferon pathway. We summarize by identifying future points of consideration that will contribute greatly to our understanding of the molecular mechanisms governing coronavirus pathogenesis and virulence, and the development of severe disease in humans and animals.
Figures

Similar articles
-
The Conserved Coronavirus Macrodomain Promotes Virulence and Suppresses the Innate Immune Response during Severe Acute Respiratory Syndrome Coronavirus Infection.mBio. 2016 Dec 13;7(6):e01721-16. doi: 10.1128/mBio.01721-16. mBio. 2016. PMID: 27965448 Free PMC article.
-
Coronavirus virulence genes with main focus on SARS-CoV envelope gene.Virus Res. 2014 Dec 19;194:124-37. doi: 10.1016/j.virusres.2014.07.024. Epub 2014 Aug 2. Virus Res. 2014. PMID: 25093995 Free PMC article. Review.
-
Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection.J Immunol. 2005 Jun 15;174(12):7977-85. doi: 10.4049/jimmunol.174.12.7977. J Immunol. 2005. PMID: 15944304
-
Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation.Microbiol Mol Biol Rev. 2008 Dec;72(4):672-85, Table of Contents. doi: 10.1128/MMBR.00015-08. Microbiol Mol Biol Rev. 2008. PMID: 19052324 Free PMC article. Review.
-
Interferon interplay helps tissue cells to cope with SARS-coronavirus infection.Virulence. 2010 Jul-Aug;1(4):273-5. doi: 10.4161/viru.1.4.11465. Virulence. 2010. PMID: 21178452
Cited by
-
Sex-related immunity: could Toll-like receptors be the answer in acute inflammatory response?Front Immunol. 2024 May 21;15:1379754. doi: 10.3389/fimmu.2024.1379754. eCollection 2024. Front Immunol. 2024. PMID: 38835761 Free PMC article. Review.
-
Analysis of Plasma Proteins Involved in Inflammation, Immune Response/Complement System, and Blood Coagulation upon Admission of COVID-19 Patients to Hospital May Help to Predict the Prognosis of the Disease.Cells. 2023 Jun 10;12(12):1601. doi: 10.3390/cells12121601. Cells. 2023. PMID: 37371071 Free PMC article.
-
Immunogenetic Role of IL17A Polymorphism in the Pathogenesis of Recurrent Miscarriage.J Clin Med. 2022 Dec 15;11(24):7448. doi: 10.3390/jcm11247448. J Clin Med. 2022. PMID: 36556060 Free PMC article.
-
HLA-C dysregulation as a possible mechanism of immune evasion in SARS-CoV-2 and other RNA-virus infections.Front Immunol. 2022 Oct 17;13:1011829. doi: 10.3389/fimmu.2022.1011829. eCollection 2022. Front Immunol. 2022. PMID: 36325330 Free PMC article.
-
Analysis of post COVID-19 condition and its overlap with myalgic encephalomyelitis/chronic fatigue syndrome.J Adv Res. 2022 Sep;40:179-196. doi: 10.1016/j.jare.2021.11.013. Epub 2021 Nov 26. J Adv Res. 2022. PMID: 36100326 Free PMC article. Review.
References
-
- Akira S., Hemmi H. Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett. 2003;85(2):85–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

