Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53
- PMID: 17438265
- PMCID: PMC1855428
- DOI: 10.1073/pnas.0702010104
Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53
Abstract
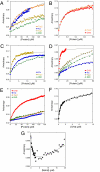
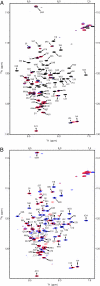
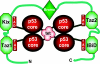
The transcriptional coactivator p300 binds to and mediates the transcriptional functions of the tetrameric tumor suppressor p53. Both proteins consist of independently folded domains linked by intrinsically disordered sequences. A well studied short sequence of the p53 transactivation domain, p53(15-29), binds weakly to four folded domains of p300 [Taz1/cysteine-histidine-rich region 1 (CH1), Kix, Taz2/CH3, IBiD], with dissociation constants (K(D)) in the 100 muM region. However, we found that a longer N-terminal transactivation domain construct p53(1-57) bound tightly to each p300 domain. Taz2/CH3 had the greatest affinity (K(D) = 27 nM) and competes with the N-terminal domain of Mdm2 for the p53 N terminus. p300 thus can protect the N terminus of p53 against the binding of other proteins. Mutations of p53 that abrogate transactivation (L22Q/W23S, W53Q/F54S) greatly weakened binding to each p300 domain, linking phenotypic defects to weakened coactivator binding. We propose a complex between tetrameric p53 and p300 in which four domains of p300 wrap around the four transactivation domains of p53.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

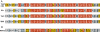
Similar articles
-
Two distinct motifs within the p53 transactivation domain bind to the Taz2 domain of p300 and are differentially affected by phosphorylation.Biochemistry. 2009 Feb 17;48(6):1244-55. doi: 10.1021/bi801716h. Biochemistry. 2009. PMID: 19166313 Free PMC article.
-
Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2.Oncogene. 2009 May 21;28(20):2112-8. doi: 10.1038/onc.2009.71. Epub 2009 Apr 13. Oncogene. 2009. PMID: 19363523 Free PMC article.
-
Molecular basis of the interactions between the p73 N terminus and p300: effects on transactivation and modulation by phosphorylation.Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3142-7. doi: 10.1073/pnas.0900383106. Epub 2009 Feb 13. Proc Natl Acad Sci U S A. 2009. PMID: 19218448 Free PMC article.
-
Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2.Proc Natl Acad Sci U S A. 2009 Apr 21;106(16):6591-6. doi: 10.1073/pnas.0811023106. Epub 2009 Apr 8. Proc Natl Acad Sci U S A. 2009. PMID: 19357310 Free PMC article.
-
Role of Intrinsic Protein Disorder in the Function and Interactions of the Transcriptional Coactivators CREB-binding Protein (CBP) and p300.J Biol Chem. 2016 Mar 25;291(13):6714-22. doi: 10.1074/jbc.R115.692020. Epub 2016 Feb 5. J Biol Chem. 2016. PMID: 26851278 Free PMC article. Review.
Cited by
-
Characterization of the p300 Taz2-p53 TAD2 complex and comparison with the p300 Taz2-p53 TAD1 complex.Biochemistry. 2015 Mar 24;54(11):2001-10. doi: 10.1021/acs.biochem.5b00044. Epub 2015 Mar 16. Biochemistry. 2015. PMID: 25753752 Free PMC article.
-
Long-range regulation of p53 DNA binding by its intrinsically disordered N-terminal transactivation domain.Proc Natl Acad Sci U S A. 2018 Nov 27;115(48):E11302-E11310. doi: 10.1073/pnas.1814051115. Epub 2018 Nov 12. Proc Natl Acad Sci U S A. 2018. PMID: 30420502 Free PMC article.
-
Activating transcription factor 3 activates p53 by preventing E6-associated protein from binding to E6.J Biol Chem. 2010 Apr 23;285(17):13201-10. doi: 10.1074/jbc.M109.058669. Epub 2010 Feb 18. J Biol Chem. 2010. PMID: 20167600 Free PMC article.
-
Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation.Proc Natl Acad Sci U S A. 2010 Nov 9;107(45):19290-5. doi: 10.1073/pnas.1013078107. Epub 2010 Oct 20. Proc Natl Acad Sci U S A. 2010. PMID: 20962272 Free PMC article.
-
The F-box protein beta-TrCp1/Fbw1a interacts with p300 to enhance beta-catenin transcriptional activity.J Biol Chem. 2009 May 8;284(19):13033-44. doi: 10.1074/jbc.M901248200. Epub 2009 Mar 17. J Biol Chem. 2009. PMID: 19297328 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

