Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling
- PMID: 17437629
- PMCID: PMC1858702
- DOI: 10.1186/1471-2199-8-28
Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling
Abstract
Background: KSRP is a AU-rich element (ARE) binding protein that causes decay of select sets of transcripts in different cell types. We have recently described that phosphatidylinositol 3-kinase/AKT (PI3K-AKT) activation induces stabilization and accumulation of the labile beta-catenin mRNA through an impairment of KSRP function.
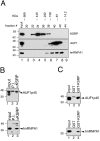
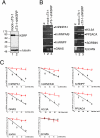
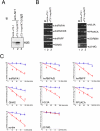
Results: Aim of this study was to identify additional KSRP targets whose stability and steady-state levels are enhanced by PI3K-AKT activation. First, through microarray analyses of the AU-rich transcriptome in pituitary alphaT3-1 cells, we identified 34 ARE-containing transcripts upregulated in cells expressing a constitutively active form of AKT1. In parallel, by an affinity chromatography-based technique followed by microarray analyses, 12 mRNAs target of KSRP, additional to beta-catenin, were identified. Among them, seven mRNAs were upregulated in cells expressing activated AKT1. Both steady-state levels and stability of these new KSRP targets were consistently increased by either KSRP knock-down or PI3K-AKT activation.
Conclusion: Our study identified a set of transcripts that are targets of KSRP and whose expression is increased by PI3K-AKT activation. These mRNAs encode RNA binding proteins, signaling molecules and a replication-independent histone. The increased expression of these gene products upon PI3K-AKT activation could play a role in the cellular events initiated by this signaling pathway.
Figures


Similar articles
-
The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling.PLoS Biol. 2006 Dec;5(1):e5. doi: 10.1371/journal.pbio.0050005. PLoS Biol. 2006. Retraction in: PLoS Biol. 2015 Nov 16;13(11):e1002314. doi: 10.1371/journal.pbio.1002314 PMID: 17177604 Free PMC article. Retracted.
-
PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis.Cell Death Differ. 2012 Mar;19(3):478-87. doi: 10.1038/cdd.2011.117. Epub 2011 Sep 2. Cell Death Differ. 2012. PMID: 21886180 Free PMC article.
-
The KH-type splicing regulatory protein (KSRP) regulates type III interferon expression post-transcriptionally.Biochem J. 2019 Jan 31;476(2):333-352. doi: 10.1042/BCJ20180522. Biochem J. 2019. PMID: 30578289
-
The role of KSRP in mRNA decay and microRNA precursor maturation.Wiley Interdiscip Rev RNA. 2010 Sep-Oct;1(2):230-9. doi: 10.1002/wrna.2. Epub 2010 May 6. Wiley Interdiscip Rev RNA. 2010. PMID: 21935887 Review.
-
Functional and molecular insights into KSRP function in mRNA decay.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):689-94. doi: 10.1016/j.bbagrm.2012.11.003. Epub 2012 Nov 22. Biochim Biophys Acta. 2013. PMID: 23178464 Review.
Cited by
-
Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence.Biol Chem. 2008 Mar;389(3):243-55. doi: 10.1515/BC.2008.022. Biol Chem. 2008. PMID: 18177264 Free PMC article. Review.
-
K-homology splicing regulatory protein (KSRP) promotes post-transcriptional destabilization of Spry4 transcripts in non-small cell lung cancer.J Biol Chem. 2017 May 5;292(18):7423-7434. doi: 10.1074/jbc.M116.757906. Epub 2017 Mar 8. J Biol Chem. 2017. PMID: 28275056 Free PMC article.
-
Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14-3-3 binding.Nat Struct Mol Biol. 2009 Mar;16(3):238-46. doi: 10.1038/nsmb.1558. Epub 2009 Feb 8. Nat Struct Mol Biol. 2009. PMID: 19198587 Free PMC article.
-
Role of KSRP in control of type I interferon and cytokine expression.J Interferon Cytokine Res. 2014 Apr;34(4):267-74. doi: 10.1089/jir.2013.0143. J Interferon Cytokine Res. 2014. PMID: 24697204 Free PMC article. Review.
-
The RNA-binding protein hnRNPA2 regulates β-catenin protein expression and is overexpressed in prostate cancer.RNA Biol. 2014;11(6):755-65. doi: 10.4161/rna.28800. Epub 2014 Apr 24. RNA Biol. 2014. PMID: 24823909 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

