US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells
- PMID: 17428859
- PMCID: PMC1900093
- DOI: 10.1128/JVI.00380-07
US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells
Abstract
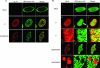
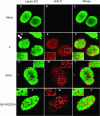
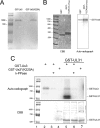
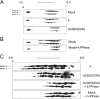
The herpes simplex virus type 1 (HSV-1) US3 gene encodes a serine/threonine kinase that, when inactivated, causes capsids to aggregate aberrantly between the inner and outer nuclear membranes (INM and ONM, respectively) within evaginations/extensions of the perinuclear space. In both Hep2 cells and an engineered cell line derived from Hep2 cells expressing lamin A/C fused to enhanced green fluorescent protein (eGFP-lamin A/C), lamin A/C localized mostly in a reticular pattern with small regions of the INM devoid of eGFP-lamin A/C when they were either mock infected or infected with wild-type HSV-1(F). Cells infected with HSV-1(F) also contained some larger diffuse regions lacking lamin A/C. Proteins UL31 and UL34, markers of potential envelopment sites at the INM and perinuclear virions, localized within the regions devoid of lamin A/C and also in regions containing lamin A/C. Similar to previous observations with Vero cells (S. L. Bjerke and R. J. Roller, Virology 347:261-276, 2006), the proteins UL34 and UL31 localized exclusively in very discrete regions of the nuclear lamina lacking lamin A/C in the absence of US3 kinase activity. To determine how US3 alters lamin A/C distribution, US3 was purified and shown to phosphorylate lamin A/C at multiple sites in vitro, despite the presence of only one putative US3 kinase consensus site in the lamin A/C sequence. US3 kinase activity was also sufficient to invoke partial solubilization of lamin A/C from permeabilized Hep2 cell nuclei in an ATP-dependent manner. Two-dimensional electrophoretic analyses of lamin A/C revealed that lamin A/C is phosphorylated in HSV-infected cells, and the full spectrum of phosphorylation requires US3 kinase activity. These data suggest that US3 kinase activity regulates HSV-1 capsid nuclear egress at least in part by phosphorylation of lamin A/C.
Figures



Similar articles
-
Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31.J Virol. 2006 Feb;80(3):1476-86. doi: 10.1128/JVI.80.3.1476-1486.2006. J Virol. 2006. PMID: 16415024 Free PMC article.
-
Effects of lamin A/C, lamin B1, and viral US3 kinase activity on viral infectivity, virion egress, and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane.J Virol. 2008 Aug;82(16):8094-104. doi: 10.1128/JVI.00874-08. Epub 2008 Jun 4. J Virol. 2008. PMID: 18524819 Free PMC article.
-
Role of herpes simplex virus 1 immediate early protein ICP22 in viral nuclear egress.J Virol. 2014 Jul;88(13):7445-54. doi: 10.1128/JVI.01057-14. Epub 2014 Apr 16. J Virol. 2014. PMID: 24741100 Free PMC article.
-
Us3 Protein Kinase Encoded by HSV: The Precise Function and Mechanism on Viral Life Cycle.Adv Exp Med Biol. 2018;1045:45-62. doi: 10.1007/978-981-10-7230-7_3. Adv Exp Med Biol. 2018. PMID: 29896662 Review.
-
[Molecular mechanism by which Us3 protein kinase regulates the pathogenicity of herpes simplex virus type-1].Uirusu. 2016;66(1):83-90. doi: 10.2222/jsv.66.83. Uirusu. 2016. PMID: 28484184 Review. Japanese.
Cited by
-
Mechanism of Nuclear Lamina Disruption and the Role of pUS3 in HSV-1 Nuclear Egress.J Virol. 2021 Apr 26;95(10):e02432-20. doi: 10.1128/JVI.02432-20. Epub 2021 Mar 3. J Virol. 2021. PMID: 33658339 Free PMC article.
-
The herpes simplex virus 2 UL21 protein is essential for virus propagation.J Virol. 2013 May;87(10):5904-15. doi: 10.1128/JVI.03489-12. Epub 2013 Mar 13. J Virol. 2013. PMID: 23487471 Free PMC article.
-
Simian virus 40 T/t antigens and lamin A/C small interfering RNA rescue the phenotype of an Epstein-Barr virus protein kinase (BGLF4) mutant.J Virol. 2010 May;84(9):4524-33. doi: 10.1128/JVI.02456-09. Epub 2010 Feb 10. J Virol. 2010. PMID: 20147387 Free PMC article.
-
Equine Herpesvirus 1 Bridles T Lymphocytes To Reach Its Target Organs.J Virol. 2019 Mar 21;93(7):e02098-18. doi: 10.1128/JVI.02098-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651370 Free PMC article.
-
Viral serine/threonine protein kinases.J Virol. 2011 Feb;85(3):1158-73. doi: 10.1128/JVI.01369-10. Epub 2010 Nov 17. J Virol. 2011. PMID: 21084474 Free PMC article. Review.
References
-
- Collas, P. 1999. Sequential PKC- and Cdc2-mediated phosphorylation events elicit zebrafish nuclear envelope disassembly. J. Cell Sci. 112:977-987. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

