Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch
- PMID: 17409150
- PMCID: PMC1900129
- DOI: 10.1128/JVI.02817-06
Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch
Abstract
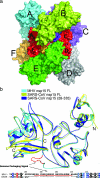
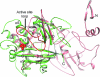
Mature nonstructural protein-15 (nsp15) from the severe acute respiratory syndrome coronavirus (SARS-CoV) contains a novel uridylate-specific Mn2+-dependent endoribonuclease (NendoU). Structure studies of the full-length form of the obligate hexameric enzyme from two CoVs, SARS-CoV and murine hepatitis virus, and its monomeric homologue, XendoU from Xenopus laevis, combined with mutagenesis studies have implicated several residues in enzymatic activity and the N-terminal domain as the major determinant of hexamerization. However, the tight link between hexamerization and enzyme activity in NendoUs has remained an enigma. Here, we report the structure of a trimmed, monomeric form of SARS-CoV nsp15 (residues 28 to 335) determined to a resolution of 2.9 A. The catalytic loop (residues 234 to 249) with its two reactive histidines (His 234 and His 249) is dramatically flipped by approximately 120 degrees into the active site cleft. Furthermore, the catalytic nucleophile Lys 289 points in a diametrically opposite direction, a consequence of an outward displacement of the supporting loop (residues 276 to 295). In the full-length hexameric forms, these two loops are packed against each other and are stabilized by intimate intersubunit interactions. Our results support the hypothesis that absence of an adjacent monomer due to deletion of the hexamerization domain is the most likely cause for disruption of the active site, offering a structural basis for why only the hexameric form of this enzyme is active.
Figures


Similar articles
-
Structural and Biochemical Characterization of Endoribonuclease Nsp15 Encoded by Middle East Respiratory Syndrome Coronavirus.J Virol. 2018 Oct 29;92(22):e00893-18. doi: 10.1128/JVI.00893-18. Print 2018 Nov 15. J Virol. 2018. PMID: 30135128 Free PMC article.
-
Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.Proc Natl Acad Sci U S A. 2006 Aug 8;103(32):11892-7. doi: 10.1073/pnas.0601708103. Epub 2006 Aug 1. Proc Natl Acad Sci U S A. 2006. PMID: 16882730 Free PMC article.
-
Insight into the evolution of nidovirus endoribonuclease based on the finding that nsp15 from porcine Deltacoronavirus functions as a dimer.J Biol Chem. 2018 Aug 3;293(31):12054-12067. doi: 10.1074/jbc.RA118.003756. Epub 2018 Jun 10. J Biol Chem. 2018. PMID: 29887523 Free PMC article.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
-
The coronavirus replicase: insights into a sophisticated enzyme machinery.Adv Exp Med Biol. 2006;581:3-11. doi: 10.1007/978-0-387-33012-9_1. Adv Exp Med Biol. 2006. PMID: 17037497 Free PMC article. Review. No abstract available.
Cited by
-
An "Old" protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses.Virology. 2018 Apr;517:157-163. doi: 10.1016/j.virol.2017.12.024. Epub 2018 Jan 4. Virology. 2018. PMID: 29307596 Free PMC article. Review.
-
Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication.PLoS One. 2008 Oct 1;3(10):e3299. doi: 10.1371/journal.pone.0003299. PLoS One. 2008. PMID: 18827877 Free PMC article.
-
SARS-CoV-2 Nsp15 suppresses type I interferon production by inhibiting IRF3 phosphorylation and nuclear translocation.iScience. 2023 Aug 23;26(9):107705. doi: 10.1016/j.isci.2023.107705. eCollection 2023 Sep 15. iScience. 2023. PMID: 37680466 Free PMC article.
-
Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling.J Virol. 2009 Oct;83(19):10314-8. doi: 10.1128/JVI.00842-09. Epub 2009 Jul 29. J Virol. 2009. PMID: 19640993 Free PMC article.
-
Cryo-EM Structures of the SARS-CoV-2 Endoribonuclease Nsp15.bioRxiv [Preprint]. 2020 Aug 11:2020.08.11.244863. doi: 10.1101/2020.08.11.244863. bioRxiv. 2020. Update in: Nat Commun. 2021 Jan 27;12(1):636. doi: 10.1038/s41467-020-20608-z. PMID: 32803198 Free PMC article. Updated. Preprint.
References
-
- Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. - PubMed
-
- Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

