Chromatin immunoprecipitation and microarray-based analysis of protein location
- PMID: 17406303
- PMCID: PMC3004291
- DOI: 10.1038/nprot.2006.98
Chromatin immunoprecipitation and microarray-based analysis of protein location
Abstract
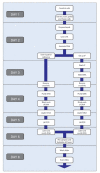
Genome-wide location analysis, also known as ChIP-Chip, combines chromatin immunoprecipitation and DNA microarray analysis to identify protein-DNA interactions that occur in living cells. Protein-DNA interactions are captured in vivo by chemical crosslinking. Cell lysis, DNA fragmentation and immunoaffinity purification of the desired protein will co-purify DNA fragments that are associated with that protein. The enriched DNA population is then labeled, combined with a differentially labeled reference sample and applied to DNA microarrays to detect enriched signals. Various computational and bioinformatic approaches are then applied to normalize the enriched and reference channels, to connect signals to the portions of the genome that are represented on the DNA microarrays, to provide confidence metrics and to generate maps of protein-genome occupancy. Here, we describe the experimental protocols that we use from crosslinking of cells to hybridization of labeled material, together with insights into the aspects of these protocols that influence the results. These protocols require approximately 1 week to complete once sufficient numbers of cells have been obtained, and have been used to produce robust, high-quality ChIP-chip results in many different cell and tissue types.
Figures



Similar articles
-
Chromatin immunoprecipitation using microarrays.Methods Mol Biol. 2009;529:279-95. doi: 10.1007/978-1-59745-538-1_18. Methods Mol Biol. 2009. PMID: 19381973
-
Chromatin Immunoprecipitation: Application to the Study of Asthma.Methods Mol Biol. 2016;1434:121-37. doi: 10.1007/978-1-4939-3652-6_9. Methods Mol Biol. 2016. PMID: 27300535
-
Mapping the distribution of chromatin proteins by ChIP on chip.Methods Enzymol. 2006;410:316-41. doi: 10.1016/S0076-6879(06)10015-4. Methods Enzymol. 2006. PMID: 16938558 Review.
-
Genome-wide mapping of protein-DNA interaction by chromatin immunoprecipitation and DNA microarray hybridization (ChIP-chip). Part A: ChIP-chip molecular methods.Methods Mol Biol. 2010;631:139-60. doi: 10.1007/978-1-60761-646-7_12. Methods Mol Biol. 2010. PMID: 20204874
-
Genomic analysis of protein-DNA interactions in bacteria: insights into transcription and chromosome organization.Mol Microbiol. 2007 Jul;65(1):21-6. doi: 10.1111/j.1365-2958.2007.05781.x. Mol Microbiol. 2007. PMID: 17581117 Review.
Cited by
-
Real-time analysis and selection of methylated DNA by fluorescence-activated single molecule sorting in a nanofluidic channel.Proc Natl Acad Sci U S A. 2012 May 29;109(22):8477-82. doi: 10.1073/pnas.1117549109. Epub 2012 May 14. Proc Natl Acad Sci U S A. 2012. PMID: 22586076 Free PMC article.
-
Kruppel-like factor 15 (KLF15) is a key regulator of podocyte differentiation.J Biol Chem. 2012 Jun 1;287(23):19122-35. doi: 10.1074/jbc.M112.345983. Epub 2012 Apr 9. J Biol Chem. 2012. PMID: 22493483 Free PMC article.
-
Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation.Nat Neurosci. 2013 Apr;16(4):434-40. doi: 10.1038/nn.3354. Epub 2013 Mar 10. Nat Neurosci. 2013. PMID: 23475113 Free PMC article.
-
Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis.Gut. 2016 Sep;65(9):1482-93. doi: 10.1136/gutjnl-2014-308614. Epub 2015 Jun 12. Gut. 2016. PMID: 26071132 Free PMC article.
-
Epigenetic marks define the lineage and differentiation potential of two distinct neural crest-derived intermediate odontogenic progenitor populations.Stem Cells Dev. 2013 Jun 15;22(12):1763-78. doi: 10.1089/scd.2012.0711. Epub 2013 Mar 15. Stem Cells Dev. 2013. PMID: 23379639 Free PMC article.
References
-
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. - PubMed
-
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. - PubMed
-
- Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell. 2000;6:1297–1307. - PubMed
-
- Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

