Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50/Rta lytic switch protein functions as a tetramer
- PMID: 17392367
- PMCID: PMC1900300
- DOI: 10.1128/JVI.00140-07
Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50/Rta lytic switch protein functions as a tetramer
Abstract
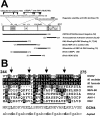
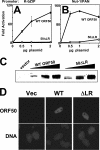
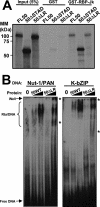
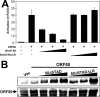
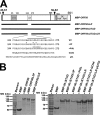
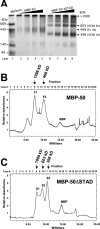
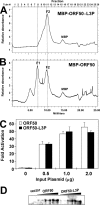
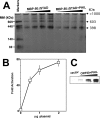
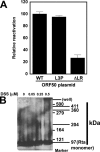
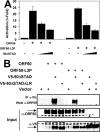
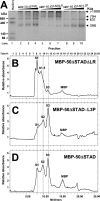
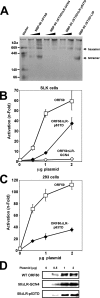
The Kaposi's sarcoma-associated herpesvirus open reading frame 50 (ORF50) protein (called Rta), is necessary and sufficient for reactivation of the virus from latency. We previously demonstrated that a truncated mutant of ORF50 lacking its C-terminal transcriptional activation domain, called ORF50DeltaSTAD, formed mixed multimers with wild-type (WT) ORF50 and functioned as a dominant negative inhibitor of reactivation. For this report, we investigated the requirements for multimerization of ORF50/Rta in transactivation and viral reactivation. We analyzed multimerization of WT, mutant, and chimeric ORF50 proteins, using Blue Native polyacrylamide gel electrophoresis and size exclusion chromatography. WT and mutant ORF50 proteins form tetramers and higher-order multimers, but not monomers, in solution. The proline-rich, N-terminal leucine heptapeptide repeat (LR) of ORF50 (amino acids [aa] 244 to 275) is necessary but not sufficient for oligomer formation and functions in concert with the central portion of ORF50/Rta (aa 245 to 414). The dominant negative mutant ORF50DeltaSTAD requires the LR to form mixed multimers with WT ORF50 and inhibit its function. In the context of the WT ORF50/Rta protein, mutagenesis of the LR, or replacement of the LR by heterologous multimerization domains from the GCN4 or p53 proteins, demonstrates that tetramers of Rta are sufficient for transactivation and viral reactivation. Mutants of Rta that are unable to form tetramers but retain the ability to form higher-order multimers are reduced in function or are nonfunctional. We concluded that the proline content, but not the leucine content, of the LR is critical for determining the oligomeric state of Rta.
Figures
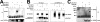
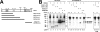
Similar articles
-
Direct interactions of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50/Rta protein with the cellular protein octamer-1 and DNA are critical for specifying transactivation of a delayed-early promoter and stimulating viral reactivation.J Virol. 2007 Aug;81(16):8451-67. doi: 10.1128/JVI.00265-07. Epub 2007 May 30. J Virol. 2007. PMID: 17537858 Free PMC article.
-
Identification of Novel Kaposi's Sarcoma-Associated Herpesvirus Orf50 Transcripts: Discovery of New RTA Isoforms with Variable Transactivation Potential.J Virol. 2016 Dec 16;91(1):e01434-16. doi: 10.1128/JVI.01434-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795414 Free PMC article.
-
Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus.Virology. 1999 Oct 25;263(2):436-49. doi: 10.1006/viro.1999.9963. Virology. 1999. PMID: 10544116
-
The Rta/Orf50 transactivator proteins of the gamma-herpesviridae.Curr Top Microbiol Immunol. 2007;312:71-100. doi: 10.1007/978-3-540-34344-8_3. Curr Top Microbiol Immunol. 2007. PMID: 17089794 Review.
-
Kaposi's sarcoma-associated herpesvirus immediate early gene activity.Front Biosci. 2004 Sep 1;9:2245-72. doi: 10.2741/1394. Front Biosci. 2004. PMID: 15353285 Review.
Cited by
-
Quantitative analysis of the bidirectional viral G-protein-coupled receptor and lytic latency-associated nuclear antigen promoter of Kaposi's sarcoma-associated herpesvirus.J Virol. 2012 Sep;86(18):9683-95. doi: 10.1128/JVI.00881-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740392 Free PMC article.
-
Genome-wide identification of binding sites for Kaposi's sarcoma-associated herpesvirus lytic switch protein, RTA.Virology. 2009 Apr 10;386(2):290-302. doi: 10.1016/j.virol.2009.01.031. Epub 2009 Feb 23. Virology. 2009. PMID: 19233445 Free PMC article.
-
A herpesvirus transactivator and cellular POU proteins extensively regulate DNA binding of the host Notch signaling protein RBP-Jκ to the virus genome.J Biol Chem. 2019 Aug 30;294(35):13073-13092. doi: 10.1074/jbc.RA118.007331. Epub 2019 Jul 15. J Biol Chem. 2019. PMID: 31308175 Free PMC article.
-
An alternative Kaposi's sarcoma-associated herpesvirus replication program triggered by host cell apoptosis.J Virol. 2012 Apr;86(8):4404-19. doi: 10.1128/JVI.06617-11. Epub 2012 Feb 15. J Virol. 2012. PMID: 22345480 Free PMC article.
-
Promoter- and cell-specific transcriptional transactivation by the Kaposi's sarcoma-associated herpesvirus ORF57/Mta protein.J Virol. 2007 Dec;81(24):13299-314. doi: 10.1128/JVI.00732-07. Epub 2007 Oct 3. J Virol. 2007. PMID: 17913801 Free PMC article.
References
-
- Ambroziak, J., D. Blackbourn, B. Herndier, R. Glogan, J. Gullet, A. McDonald, E. Lennette, and J. Levy. 1995. Herpesvirus-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268:582-583. - PubMed
-
- Baker, S. J., S. Markowitz, E. R. Fearon, J. K. Willson, and B. Vogelstein. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912-915. - PubMed
-
- Barlow, D. J., and J. M. Thornton. 1988. Helix geometry in proteins. J. Mol. Biol. 201:601-619. - PubMed
-
- Boneschi, V., L. Brambilla, E. Berti, S. Ferrucci, M. Corbellino, C. Parravicini, and S. Fossati. 2001. Human herpesvirus 8 DNA in the skin and blood of patients with Mediterranean Kaposi's sarcoma: clinical correlations. Dermatology 203:19-23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

