CMV pp65 and IE-1 T cell epitopes recognized by healthy subjects
- PMID: 17391521
- PMCID: PMC1851947
- DOI: 10.1186/1479-5876-5-17
CMV pp65 and IE-1 T cell epitopes recognized by healthy subjects
Abstract
Background: Adoptive immune and vaccine therapies have been used to prevent cytomegalovirus (CMV) disease in recipients of hematopoietic progenitor cell transplants, but the nature of T cell responses to CMV have not been completely characterized.
Methods: Peptide pools and individual peptides derived from the immune-dominant CMV proteins pp65 and IE-1 and antigen-specific, cytokine flow cytometry were used to characterize the prevalence and frequency of CD4+ and CD8+ memory T cells in 20 healthy CMV-seropositive subjects.
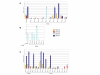
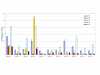
Results: CD8+ T cell responses to pp65 were detected in 35% of subjects and to IE-1 in 40% of subjects. CD4+ T cell responses to pp65 were detected in 50% of subjects, but none were detected to IE-1. Several new IE-1 HLA class I epitopes were identified, including 4 restricted to HLA-C antigens. One region of IE-1 spanning amino acids 300 to 327 was rich in class I epitopes. The HLA class I restrictions of IE-1 peptides were more promiscuous than those of pp65 peptides.
Conclusion: Since naturally occurring CD4+ and CD8+ T cell responses to pp65 were detectable in many subjects, but only CD8+ T cell responses to IE-1 were detected, pp65 may be better than IE-1 for use in vaccine and adoptive immune therapies.
Figures





Similar articles
-
Monitoring cytomegalovirus IE-1 and pp65-specific CD4+ and CD8+ T-cell responses after allogeneic stem cell transplantation may identify patients at risk for recurrent CMV reactivations.Cytometry B Clin Cytom. 2008 Jul;74(4):211-20. doi: 10.1002/cyto.b.20420. Cytometry B Clin Cytom. 2008. PMID: 18454493
-
Cytomegalovirus immune reconstitution occurs in recipients of allogeneic hematopoietic cell transplants irrespective of detectable cytomegalovirus infection.Biol Blood Marrow Transplant. 2005 Nov;11(11):890-902. doi: 10.1016/j.bbmt.2005.07.008. Biol Blood Marrow Transplant. 2005. PMID: 16275592
-
Co-ordinated isolation of CD8(+) and CD4(+) T cells recognizing a broad repertoire of cytomegalovirus pp65 and IE1 epitopes for highly specific adoptive immunotherapy.Cytotherapy. 2010 Nov;12(7):933-44. doi: 10.3109/14653240903505822. Cytotherapy. 2010. PMID: 20078388
-
Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals.J Infect Dis. 2002 Jun 15;185(12):1709-16. doi: 10.1086/340637. Epub 2002 May 31. J Infect Dis. 2002. PMID: 12085315
-
How frequently are predicted peptides actually recognized by CD8 cells?Cancer Immunol Immunother. 2016 Jul;65(7):847-55. doi: 10.1007/s00262-016-1840-7. Epub 2016 Apr 23. Cancer Immunol Immunother. 2016. PMID: 27108305 Free PMC article. Review.
Cited by
-
Identification and HLA-tetramer-validation of human CD4+ and CD8+ T cell responses against HCMV proteins IE1 and IE2.PLoS One. 2014 Apr 23;9(4):e94892. doi: 10.1371/journal.pone.0094892. eCollection 2014. PLoS One. 2014. PMID: 24760079 Free PMC article.
-
The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B*41 allotypic family.Haematologica. 2011 Jan;96(1):110-8. doi: 10.3324/haematol.2010.030924. Epub 2010 Oct 7. Haematologica. 2011. PMID: 20934997 Free PMC article.
-
Design of cocktail peptide vaccine against Cytomegalovirus infection.Iran J Basic Med Sci. 2016 Apr;19(4):449-54. Iran J Basic Med Sci. 2016. PMID: 27279990 Free PMC article.
-
Analysis of vaccine-induced T cells in humans with cancer.Adv Exp Med Biol. 2010;684:178-88. doi: 10.1007/978-1-4419-6451-9_14. Adv Exp Med Biol. 2010. PMID: 20795549 Free PMC article.
-
In-depth summary over cytomegalovirus infection in allogeneic hematopoietic stem cell transplantation recipients.Virusdisease. 2021 Sep;32(3):422-434. doi: 10.1007/s13337-021-00728-w. Epub 2021 Jul 28. Virusdisease. 2021. PMID: 34631973 Free PMC article. Review.
References
-
- Mocarski ES, Tan C. Cytomegaloviruses and their replication. In: Knipe DM and Howley PM, editor. Fields Virology. fourth. Vol. 76. Philadelphia, Lippincott Williams and Wilkins; 2001. pp. 2629–2673.
-
- Pass RF. Cytomegalovirus. In: Knipe DM and Howley PM, editor. Fields Virology. fourth. Vol. 77. Philadelphia, Lippincott Williams and Wilkins; 2001. pp. 2675–2705.
-
- Boeckh M, Leisenring W, Riddell SR, Bowden RA, Huang ML, Myerson D, Stevens-Ayers T, Flowers ME, Cunningham T, Corey L. Late cytomegalovirus disease and mortality in recipients of allogeneic hematopoietic stem cell transplants: importance of viral load and T-cell immunity. Blood. 2003;101:407–414. doi: 10.1182/blood-2002-03-0993. - DOI - PubMed
-
- Zaia JA, Sissons JG, Riddell S, Diamond DJ, Wills MR, Carmichael AJ, Weekes MP, Gandhi M, La Rosa C, Villacres M, Lacey S, Markel S, Sun J. Status of Cytomegalovirus Prevention and Treatment in 2000. Hematology (Am Soc Hematol Educ Program ) 2000:339–355. - PubMed
-
- Walter EA, Greenberg PD, Gilbert MJ, Finch RJ, Watanabe KS, Thomas ED, Riddell SR. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

