Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus
- PMID: 17389919
- PMCID: PMC1828621
- DOI: 10.1371/journal.pone.0000328
Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus
Abstract
Acanthamoeba polyphaga Mimivirus is a giant double-stranded DNA virus defining a new genus, the Mimiviridae, among the Nucleo-Cytoplasmic Large DNA Viruses (NCLDV). We used utrastructural studies to shed light on the different steps of the Mimivirus replication cycle: entry via phagocytosis, release of viral DNA into the cell cytoplasm through fusion of viral and vacuolar membranes, and finally viral morphogenesis in an extraordinary giant cytoplasmic virus factory (VF). Fluorescent staining of the AT-rich Mimivirus DNA showed that it enters the host nucleus prior to the generation of a cytoplasmic independent replication centre that forms the core of the VF. Assembly and filling of viral capsids were observed within the replication centre, before release into the cell cytoplasm where progeny virions accumulated. 3D reconstruction from fluorescent and differential contrast interference images revealed the VF emerging from the cell surface as a volcano-like structure. Its size dramatically grew during the 24 h infectious lytic cycle. Our results showed that Mimivirus replication is an extremely efficient process that results from a rapid takeover of cellular machinery, and takes place in a unique and autonomous giant assembly centre, leading to the release of a large number of complex virions through amoebal lysis.
Conflict of interest statement
Figures
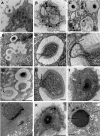

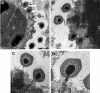

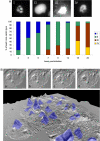
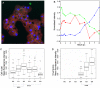
Similar articles
-
Sputnik, a virophage infecting the viral domain of life.Adv Virus Res. 2012;82:63-89. doi: 10.1016/B978-0-12-394621-8.00013-3. Adv Virus Res. 2012. PMID: 22420851 Review.
-
Infection cycles of large DNA viruses: emerging themes and underlying questions.Virology. 2014 Oct;466-467:3-14. doi: 10.1016/j.virol.2014.05.037. Epub 2014 Jul 2. Virology. 2014. PMID: 24996494 Review.
-
Complex Membrane Remodeling during Virion Assembly of the 30,000-Year-Old Mollivirus Sibericum.J Virol. 2019 Jun 14;93(13):e00388-19. doi: 10.1128/JVI.00388-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996095 Free PMC article.
-
Analyses of the Kroon Virus Major Capsid Gene and Its Transcript Highlight a Distinct Pattern of Gene Evolution and Splicing among Mimiviruses.J Virol. 2018 Jan 2;92(2):e01782-17. doi: 10.1128/JVI.01782-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118120 Free PMC article.
-
First isolation of Mimivirus in a patient with pneumonia.Clin Infect Dis. 2013 Aug;57(4):e127-34. doi: 10.1093/cid/cit354. Epub 2013 May 24. Clin Infect Dis. 2013. PMID: 23709652
Cited by
-
Membrane assembly during the infection cycle of the giant Mimivirus.PLoS Pathog. 2013;9(5):e1003367. doi: 10.1371/journal.ppat.1003367. Epub 2013 May 30. PLoS Pathog. 2013. PMID: 23737745 Free PMC article.
-
Morphogenesis of mimivirus and its viral factories: an atomic force microscopy study of infected cells.J Virol. 2013 Oct;87(20):11200-13. doi: 10.1128/JVI.01372-13. Epub 2013 Aug 7. J Virol. 2013. PMID: 23926353 Free PMC article.
-
The polyadenylation site of Mimivirus transcripts obeys a stringent 'hairpin rule'.Genome Res. 2009 Jul;19(7):1233-42. doi: 10.1101/gr.091561.109. Epub 2009 Apr 29. Genome Res. 2009. PMID: 19403753 Free PMC article.
-
Microscopic Analysis of the Tupanvirus Cycle in Vermamoeba vermiformis.Front Microbiol. 2019 Apr 3;10:671. doi: 10.3389/fmicb.2019.00671. eCollection 2019. Front Microbiol. 2019. PMID: 31001237 Free PMC article.
-
Investigating the Concept and Origin of Viruses.Trends Microbiol. 2020 Dec;28(12):959-967. doi: 10.1016/j.tim.2020.08.003. Trends Microbiol. 2020. PMID: 33158732 Free PMC article. Review.
References
-
- La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, et al. A giant virus in amoebae. Science. 2003;299:2033. - PubMed
-
- Suzan-Monti M, La Scola B, Raoult D. Genomic and evolutionary aspects of Mimivirus. Virus Res. 2006;117:145–155. - PubMed
-
- Xiao C, Chipman PR, Battisti AJ, Bowman VD, Renesto P, et al. Cryo-electron microscopy of the giant Mimivirus. J Mol Biol. 2005;353:493–496. - PubMed
-
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

