Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis
- PMID: 17384170
- PMCID: PMC1867362
- DOI: 10.1105/tpc.106.045724
Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis
Abstract
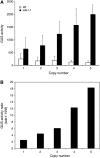
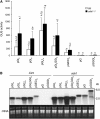
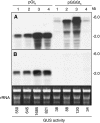
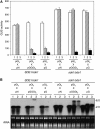
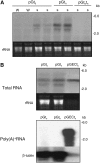
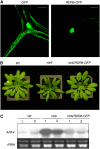
RNA silencing can be induced by highly transcribed transgenes through a pathway dependent on RNA-DEPENDENT RNA POLYMERASE6 (RDR6) and may function as a genome protection mechanism against excessively expressed genes. Whether all transcripts or just aberrant transcripts activate this protection mechanism is unclear. Consistent RNA silencing induced by a transgene with three direct repeats of the beta-glucuronidase (GUS) open reading frame (ORF) is associated with high levels of truncated, unpolyadenylated transcripts, probably from abortive transcription elongation. Truncated, unpolyadenylated transcripts from triple GUS ORF repeats were degraded in the wild type but accumulated in an rdr6 mutant, suggesting targeting for degradation by RDR6-mediated RNA silencing. A GUS transgene without a 3' transcription terminator produced unpolyadenylated readthrough mRNA and consistent RDR6-dependent RNA silencing. Both GUS triple repeats and terminator-less GUS transgenes silenced an expressed GUS transgene in trans in the wild type but not in the rdr6 mutant. Placing two 3' terminators in the GUS transgene 3' reduced mRNA 3' readthrough, decreased GUS-specific small interfering RNA accumulation, and enhanced GUS gene expression. Moreover, RDR6 was localized in the nucleus. We propose that improperly terminated, unpolyadenylated mRNA from transgene transcription is subject to RDR6-mediated RNA silencing, probably by acting as templates for the RNA polymerase, in Arabidopsis thaliana.
Figures

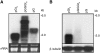

Similar articles
-
The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis.Plant J. 2012 Nov;72(3):368-77. doi: 10.1111/j.1365-313X.2012.05066.x. Epub 2012 Aug 30. Plant J. 2012. PMID: 22639932
-
High-efficiency silencing of a beta-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation.Plant Mol Biol. 2000 May;43(1):67-82. doi: 10.1023/a:1006490331303. Plant Mol Biol. 2000. PMID: 10949375
-
Silencing of invertedly repeated transgenes in Arabidopsis thaliana.Meded Rijksuniv Gent Fak Landbouwkd Toegep Biol Wet. 2001;66(3b):393-9. Meded Rijksuniv Gent Fak Landbouwkd Toegep Biol Wet. 2001. PMID: 15954624
-
Interconnections between mRNA degradation and RDR-dependent siRNA production in mRNA turnover in plants.J Plant Res. 2017 Mar;130(2):211-226. doi: 10.1007/s10265-017-0906-8. Epub 2017 Feb 14. J Plant Res. 2017. PMID: 28197782 Review.
-
Plant terminators: the unsung heroes of gene expression.J Exp Bot. 2023 Apr 9;74(7):2239-2250. doi: 10.1093/jxb/erac467. J Exp Bot. 2023. PMID: 36477559 Free PMC article. Review.
Cited by
-
Ribosome stalling and SGS3 phase separation prime the epigenetic silencing of transposons.Nat Plants. 2021 Mar;7(3):303-309. doi: 10.1038/s41477-021-00867-4. Epub 2021 Mar 1. Nat Plants. 2021. PMID: 33649597
-
Multiple actin isotypes in plants: diverse genes for diverse roles?Front Plant Sci. 2012 Oct 12;3:226. doi: 10.3389/fpls.2012.00226. eCollection 2012. Front Plant Sci. 2012. PMID: 23091476 Free PMC article.
-
A combinatorial bidirectional and bicistronic approach for coordinated multi-gene expression in corn.Plant Mol Biol. 2015 Mar;87(4-5):341-53. doi: 10.1007/s11103-015-0281-6. Epub 2015 Feb 6. Plant Mol Biol. 2015. PMID: 25657118
-
Nanovector-mediated exogenous delivery of dsRNA induces silencing of target genes in very young tomato flower buds.Nanoscale Adv. 2022 Sep 14;4(21):4542-4553. doi: 10.1039/d2na00478j. eCollection 2022 Oct 25. Nanoscale Adv. 2022. PMID: 36341284 Free PMC article.
-
Phytohormone abscisic acid control RNA-dependent RNA polymerase 6 gene expression and post-transcriptional gene silencing in rice cells.Nucleic Acids Res. 2008 Mar;36(4):1220-6. doi: 10.1093/nar/gkm1133. Epub 2007 Dec 26. Nucleic Acids Res. 2008. PMID: 18160413 Free PMC article.
References
-
- Adamson, T.E., Shutt, D.C., and Price, D.H. (2005). Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 280 32262–32271. - PubMed
-
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. - PubMed
-
- Baker, K.E., and Parker, R. (2004). Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr. Opin. Cell Biol. 16 293–299. - PubMed
-
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. - PubMed
-
- Beclin, C., Boutet, S., Waterhouse, P., and Vaucheret, H. (2002). A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12 684–688. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

