Lipid A modification systems in gram-negative bacteria
- PMID: 17362200
- PMCID: PMC2569861
- DOI: 10.1146/annurev.biochem.76.010307.145803
Lipid A modification systems in gram-negative bacteria
Abstract
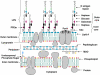
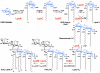
The lipid A moiety of lipopolysaccharide forms the outer monolayer of the outer membrane of most gram-negative bacteria. Escherichia coli lipid A is synthesized on the cytoplasmic surface of the inner membrane by a conserved pathway of nine constitutive enzymes. Following attachment of the core oligosaccharide, nascent core-lipid A is flipped to the outer surface of the inner membrane by the ABC transporter MsbA, where the O-antigen polymer is attached. Diverse covalent modifications of the lipid A moiety may occur during its transit from the outer surface of the inner membrane to the outer membrane. Lipid A modification enzymes are reporters for lipopolysaccharide trafficking within the bacterial envelope. Modification systems are variable and often regulated by environmental conditions. Although not required for growth, the modification enzymes modulate virulence of some gram-negative pathogens. Heterologous expression of lipid A modification enzymes may enable the development of new vaccines.
Figures
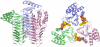
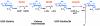




Similar articles
-
[Kdo2-lipid A modification in gram-negative bacteria--a review].Wei Sheng Wu Xue Bao. 2013 Feb 4;53(2):111-7. Wei Sheng Wu Xue Bao. 2013. PMID: 23627103 Review. Chinese.
-
Biosynthesis, transport, and modification of lipid A.Biochem Cell Biol. 2004 Feb;82(1):71-86. doi: 10.1139/o03-070. Biochem Cell Biol. 2004. PMID: 15052329 Review.
-
Discovery of new biosynthetic pathways: the lipid A story.J Lipid Res. 2009 Apr;50 Suppl(Suppl):S103-8. doi: 10.1194/jlr.R800060-JLR200. Epub 2008 Oct 29. J Lipid Res. 2009. PMID: 18974037 Free PMC article.
-
Modulation of the surface architecture of gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length.Mol Microbiol. 1997 Feb;23(4):629-38. doi: 10.1046/j.1365-2958.1997.2571614.x. Mol Microbiol. 1997. PMID: 9157235 Review.
-
[Structure of lipopolysaccharides from gram-negative bacteria. I. Common characteristics of lipopolysaccharides and lipid A structure (Review)].Biokhimiia. 1993 Feb;58(2):166-81. Biokhimiia. 1993. PMID: 8485211 Review. Russian.
Cited by
-
A novel PhoPQ-potentiated mechanism of colistin resistance impairs membrane integrity in Pseudomonas aeruginosa.bioRxiv [Preprint]. 2024 Oct 16:2024.10.15.618514. doi: 10.1101/2024.10.15.618514. bioRxiv. 2024. PMID: 39464160 Free PMC article. Preprint.
-
Elucidation of the 3-O-deacylase gene, pagL, required for the removal of primary β-hydroxy fatty acid from the lipid A in the nitrogen-fixing endosymbiont Rhizobium etli CE3.J Biol Chem. 2013 Apr 26;288(17):12004-13. doi: 10.1074/jbc.M113.470484. Epub 2013 Mar 19. J Biol Chem. 2013. PMID: 23511636 Free PMC article.
-
Characterization of galacturonosyl transferase genes rgtA, rgtB, rgtC, rgtD, and rgtE responsible for lipopolysaccharide synthesis in nitrogen-fixing endosymbiont Rhizobium leguminosarum: lipopolysaccharide core and lipid galacturonosyl residues confer membrane stability.J Biol Chem. 2012 Jan 6;287(2):935-49. doi: 10.1074/jbc.M111.311571. Epub 2011 Nov 22. J Biol Chem. 2012. PMID: 22110131 Free PMC article.
-
More than Meets the Eye: Untargeted Metabolomics and Lipidomics Reveal Complex Pathways Spurred by Activation of Acid Resistance Mechanisms in Escherichia coli.J Proteome Res. 2022 Dec 2;21(12):2958-2968. doi: 10.1021/acs.jproteome.2c00459. Epub 2022 Nov 2. J Proteome Res. 2022. PMID: 36322795 Free PMC article.
-
Membrane Fractionation by Isopycnic Sucrose Density Gradient Centrifugation for Qualitative Analysis of LPS in Escherichia coli.Methods Mol Biol. 2022;2548:53-69. doi: 10.1007/978-1-0716-2581-1_4. Methods Mol Biol. 2022. PMID: 36151491
References
-
- Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr., et al. J. Lipid Res. 2005;46:839–62. - PubMed
-
- Brade H, Opal SM, Vogel SN, Morrison DC, editors. Endotoxin in Health and Disease. Marcel Dekker, Inc.; New York: 1999. p. 950.
-
- Galloway SM, Raetz CRH. J. Biol. Chem. 1990;265:6394–402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

