Global extent of horizontal gene transfer
- PMID: 17360551
- PMCID: PMC1815472
- DOI: 10.1073/pnas.0611557104
Global extent of horizontal gene transfer
Abstract
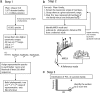
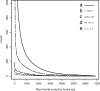
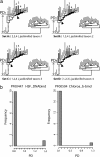
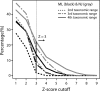
Horizontal gene transfer (HGT) is thought to play an important role in the evolution of species and innovation of genomes. There have been many convincing evidences for HGT for specific genes or gene families, but there has been no estimate of the global extent of HGT. Here, we present a method of identifying HGT events within a given protein family and estimate the global extent of HGT in all curated protein domain families ( approximately 8,000) listed in the Pfam database. The results suggest four conclusions: (i) for all protein domain families in Pfam, the fixation of genes horizontally transferred is not a rampant phenomenon between organisms with substantial phylogenetic separations (1.1-9.7% of Pfam families surveyed at three taxonomic ranges studied show indication of HGT); (ii) however, at the level of domains, >50% of Archaea have one or more protein domains acquired by HGT, and nearly 30-50% of Bacteria did the same when examined at three taxonomic ranges. But, the equivalent value for Eukarya is <10%; (iii) HGT will have very little impact in the construction of organism phylogeny, when the construction methods use whole genomes, large numbers of common genes, or SSU rRNAs; and (iv) there appears to be no strong preference of HGT for protein families of particular cellular or molecular functions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
A database of phylogenetically atypical genes in archaeal and bacterial genomes, identified using the DarkHorse algorithm.BMC Bioinformatics. 2008 Oct 7;9:419. doi: 10.1186/1471-2105-9-419. BMC Bioinformatics. 2008. PMID: 18840280 Free PMC article.
-
Identification of horizontally transferred genes in the genus Colletotrichum reveals a steady tempo of bacterial to fungal gene transfer.BMC Genomics. 2015 Jan 2;16(1):2. doi: 10.1186/1471-2164-16-2. BMC Genomics. 2015. PMID: 25555398 Free PMC article.
-
The cobweb of life revealed by genome-scale estimates of horizontal gene transfer.PLoS Biol. 2005 Oct;3(10):e316. doi: 10.1371/journal.pbio.0030316. Epub 2005 Aug 30. PLoS Biol. 2005. PMID: 16122348 Free PMC article.
-
Horizontal gene transfer: a critical view.Proc Natl Acad Sci U S A. 2003 Aug 19;100(17):9658-62. doi: 10.1073/pnas.1632870100. Epub 2003 Aug 5. Proc Natl Acad Sci U S A. 2003. PMID: 12902542 Free PMC article. Review.
-
Horizontal gene transfer in fungi.FEMS Microbiol Lett. 2012 Apr;329(1):1-8. doi: 10.1111/j.1574-6968.2011.02465.x. Epub 2011 Dec 15. FEMS Microbiol Lett. 2012. PMID: 22112233 Review.
Cited by
-
The Astrobiology Primer v2.0.Astrobiology. 2016 Aug;16(8):561-653. doi: 10.1089/ast.2015.1460. Astrobiology. 2016. PMID: 27532777 Free PMC article. Review. No abstract available.
-
Phylogenomic analysis identifies gene gains that define Salmonella enterica subspecies I.PLoS One. 2013 Oct 28;8(10):e76821. doi: 10.1371/journal.pone.0076821. eCollection 2013. PLoS One. 2013. PMID: 24204679 Free PMC article.
-
Activation of SsoPK4, an Archaeal eIF2α Kinase Homolog, by Oxidized CoA.Proteomes. 2015 May 15;3(2):89-116. doi: 10.3390/proteomes3020089. Proteomes. 2015. PMID: 28248264 Free PMC article.
-
Predicting plasmid promiscuity based on genomic signature.J Bacteriol. 2010 Nov;192(22):6045-55. doi: 10.1128/JB.00277-10. Epub 2010 Sep 17. J Bacteriol. 2010. PMID: 20851899 Free PMC article.
-
Benefits of using molecular structure and abundance in phylogenomic analysis.Front Genet. 2012 Sep 6;3:172. doi: 10.3389/fgene.2012.00172. eCollection 2012. Front Genet. 2012. PMID: 22973296 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

