Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity
- PMID: 17360516
- PMCID: PMC1810507
- DOI: 10.1073/pnas.0603762104
Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity
Abstract
Histone chaperones assemble and disassemble nucleosomes in an ATP-independent manner and thus regulate the most fundamental step in the alteration of chromatin structure. The molecular mechanisms underlying histone chaperone activity remain unclear. To gain insights into these mechanisms, we solved the crystal structure of the functional domain of SET/TAF-Ibeta/INHAT at a resolution of 2.3 A. We found that SET/TAF-Ibeta/INHAT formed a dimer that assumed a "headphone"-like structure. Each subunit of the SET/TAF-Ibeta/INHAT dimer consisted of an N terminus, a backbone helix, and an "earmuff" domain. It resembles the structure of the related protein NAP-1. Comparison of the crystal structures of SET/TAF-Ibeta/INHAT and NAP-1 revealed that the two proteins were folded similarly except for an inserted helix. However, their backbone helices were shaped differently, and the relative dispositions of the backbone helix and the earmuff domain between the two proteins differed by approximately 40 degrees . Our biochemical analyses of mutants revealed that the region of SET/TAF-Ibeta/INHAT that is engaged in histone chaperone activity is the bottom surface of the earmuff domain, because this surface bound both core histones and double-stranded DNA. This overlap or closeness of the activity surface and the binding surfaces suggests that the specific association among SET/TAF-Ibeta/INHAT, core histones, and double-stranded DNA is requisite for histone chaperone activity. These findings provide insights into the possible mechanisms by which histone chaperones assemble and disassemble nucleosome structures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

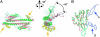

Similar articles
-
Identification of distinct SET/TAF-Ibeta domains required for core histone binding and quantitative characterisation of the interaction.BMC Biochem. 2009 Apr 9;10:10. doi: 10.1186/1471-2091-10-10. BMC Biochem. 2009. PMID: 19358706 Free PMC article.
-
A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Ibeta in integrating chromatin hypoacetylation and transcriptional repression.J Biol Chem. 2004 Jul 16;279(29):30850-5. doi: 10.1074/jbc.M404969200. Epub 2004 May 10. J Biol Chem. 2004. PMID: 15136563
-
Effect of leucine-to-methionine substitutions on the diffraction quality of histone chaperone SET/TAF-Ibeta/INHAT crystals.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 Oct 1;64(Pt 10):960-5. doi: 10.1107/S1744309108028704. Epub 2008 Sep 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18931446 Free PMC article.
-
Histone chaperones and nucleosome assembly.Curr Opin Struct Biol. 2003 Feb;13(1):6-14. doi: 10.1016/s0959-440x(03)00002-2. Curr Opin Struct Biol. 2003. PMID: 12581654 Review.
-
Regulation of chromatin structure and function: insights into the histone chaperone FACT.Cell Cycle. 2021 Mar-Mar;20(5-6):465-479. doi: 10.1080/15384101.2021.1881726. Epub 2021 Feb 16. Cell Cycle. 2021. PMID: 33590780 Free PMC article. Review.
Cited by
-
Regulation of SET Gene Expression by NFkB.Mol Neurobiol. 2017 Aug;54(6):4477-4485. doi: 10.1007/s12035-016-9967-2. Epub 2016 Jun 28. Mol Neurobiol. 2017. PMID: 27351675
-
Mass spectrometry-based multi-omics analysis reveals distinct molecular features in early and advanced stages of hepatocellular carcinoma.Heliyon. 2024 Sep 20;10(19):e38182. doi: 10.1016/j.heliyon.2024.e38182. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39381095 Free PMC article.
-
Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading.Mol Cell. 2016 Oct 6;64(1):79-91. doi: 10.1016/j.molcel.2016.08.022. Epub 2016 Sep 22. Mol Cell. 2016. PMID: 27666591 Free PMC article.
-
Thirty years of SET/TAF1β/I2PP2A: from the identification of the biological functions to its implications in cancer and Alzheimer's disease.Biosci Rep. 2022 Nov 30;42(11):BSR20221280. doi: 10.1042/BSR20221280. Biosci Rep. 2022. PMID: 36345878 Free PMC article. Review.
-
Mitochondrial cytochrome c shot towards histone chaperone condensates in the nucleus.FEBS Open Bio. 2021 Sep;11(9):2418-2440. doi: 10.1002/2211-5463.13176. Epub 2021 May 19. FEBS Open Bio. 2021. PMID: 33938164 Free PMC article. Review.
References
-
- Kornberg RD. Science. 1974;184:868–871. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. - PubMed
-
- Jenuwein T, Allis CD. Science. 2001;293:1074–1080. - PubMed
-
- Mellor J. Mol Cell. 2005;19:147–157. - PubMed
-
- Horn PJ, Peterson CL. Science. 2002;297:1824–1827. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

