Large-scale mapping of human protein-protein interactions by mass spectrometry
- PMID: 17353931
- PMCID: PMC1847948
- DOI: 10.1038/msb4100134
Large-scale mapping of human protein-protein interactions by mass spectrometry
Abstract
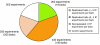
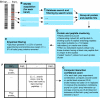
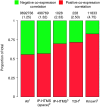
Mapping protein-protein interactions is an invaluable tool for understanding protein function. Here, we report the first large-scale study of protein-protein interactions in human cells using a mass spectrometry-based approach. The study maps protein interactions for 338 bait proteins that were selected based on known or suspected disease and functional associations. Large-scale immunoprecipitation of Flag-tagged versions of these proteins followed by LC-ESI-MS/MS analysis resulted in the identification of 24,540 potential protein interactions. False positives and redundant hits were filtered out using empirical criteria and a calculated interaction confidence score, producing a data set of 6463 interactions between 2235 distinct proteins. This data set was further cross-validated using previously published and predicted human protein interactions. In-depth mining of the data set shows that it represents a valuable source of novel protein-protein interactions with relevance to human diseases. In addition, via our preliminary analysis, we report many novel protein interactions and pathway associations.
Figures

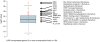

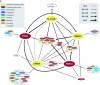
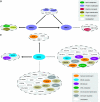
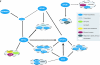
Similar articles
-
Quantifying carbohydrate-protein interactions by electrospray ionization mass spectrometry analysis.Biochemistry. 2012 May 29;51(21):4244-53. doi: 10.1021/bi300436x. Epub 2012 May 17. Biochemistry. 2012. PMID: 22564104
-
In-spray supercharging of peptides and proteins in electrospray ionization mass spectrometry.Anal Chem. 2012 Jun 5;84(11):4647-51. doi: 10.1021/ac300845n. Epub 2012 May 11. Anal Chem. 2012. PMID: 22571167
-
Reactive-electrospray-assisted laser desorption/ionization for characterization of peptides and proteins.Anal Chem. 2008 Sep 15;80(18):6995-7003. doi: 10.1021/ac800870c. Epub 2008 Aug 7. Anal Chem. 2008. PMID: 18683952
-
Electrospray-ionization mass spectrometry as a tool for fast screening of protein structural properties.Biotechnol J. 2009 Jan;4(1):73-87. doi: 10.1002/biot.200800250. Biotechnol J. 2009. PMID: 19156745 Review.
-
Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins.Mass Spectrom Rev. 2001 Mar-Apr;20(2):61-87. doi: 10.1002/mas.1003. Mass Spectrom Rev. 2001. PMID: 11455562 Review.
Cited by
-
A human skeletal muscle interactome centered on proteins involved in muscular dystrophies: LGMD interactome.Skelet Muscle. 2013 Feb 15;3(1):3. doi: 10.1186/2044-5040-3-3. Skelet Muscle. 2013. PMID: 23414517 Free PMC article.
-
Proteomic analysis of cellular soluble proteins from human bronchial smooth muscle cells by combining nondenaturing micro 2DE and quantitative LC-MS/MS. 2. Similarity search between protein maps for the analysis of protein complexes.Electrophoresis. 2015 Sep;36(17):1991-2001. doi: 10.1002/elps.201400574. Epub 2015 Jul 20. Electrophoresis. 2015. PMID: 26031785 Free PMC article.
-
Non Linear Programming (NLP) formulation for quantitative modeling of protein signal transduction pathways.PLoS One. 2012;7(11):e50085. doi: 10.1371/journal.pone.0050085. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226239 Free PMC article.
-
A protein-protein interaction analysis tool for targeted cross-linking mass spectrometry.Sci Rep. 2023 Dec 13;13(1):22103. doi: 10.1038/s41598-023-49663-4. Sci Rep. 2023. PMID: 38092875 Free PMC article.
-
Carboxypeptidase E, Identified As a Direct Interactor of Growth Hormone, Is Important for Efficient Secretion of the Hormone.Mol Cells. 2016 Oct;39(10):756-761. doi: 10.14348/molcells.2016.0183. Epub 2016 Oct 28. Mol Cells. 2016. PMID: 27788574 Free PMC article.
References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 - PMC - PubMed
-
- Bader GD, Hogue CW (2002) Analyzing yeast protein–protein interaction data obtained from different sources. Nat Biotechnol 20: 991–997 - PubMed
-
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL (2005) High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307: 1621–1625 - PubMed
-
- Belham C, Roig J, Caldwell JA, Aoyama Y, Kemp BE, Comb M, Avruch J (2003) A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J Biol Chem 278: 34897–34909 - PubMed
-
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

