Essential gene identification and drug target prioritization in Aspergillus fumigatus
- PMID: 17352532
- PMCID: PMC1817658
- DOI: 10.1371/journal.ppat.0030024
Essential gene identification and drug target prioritization in Aspergillus fumigatus
Abstract
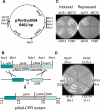
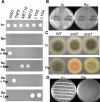
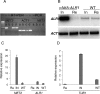
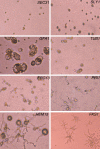
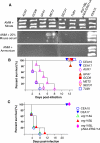
Aspergillus fumigatus is the most prevalent airborne filamentous fungal pathogen in humans, causing severe and often fatal invasive infections in immunocompromised patients. Currently available antifungal drugs to treat invasive aspergillosis have limited modes of action, and few are safe and effective. To identify and prioritize antifungal drug targets, we have developed a conditional promoter replacement (CPR) strategy using the nitrogen-regulated A. fumigatus NiiA promoter (pNiiA). The gene essentiality for 35 A. fumigatus genes was directly demonstrated by this pNiiA-CPR strategy from a set of 54 genes representing broad biological functions whose orthologs are confirmed to be essential for growth in Candida albicans and Saccharomyces cerevisiae. Extending this approach, we show that the ERG11 gene family (ERG11A and ERG11B) is essential in A. fumigatus despite neither member being essential individually. In addition, we demonstrate the pNiiA-CPR strategy is suitable for in vivo phenotypic analyses, as a number of conditional mutants, including an ERG11 double mutant (erg11BDelta, pNiiA-ERG11A), failed to establish a terminal infection in an immunocompromised mouse model of systemic aspergillosis. Collectively, the pNiiA-CPR strategy enables a rapid and reliable means to directly identify, phenotypically characterize, and facilitate target-based whole cell assays to screen A. fumigatus essential genes for cognate antifungal inhibitors.
Conflict of interest statement
Figures


Similar articles
-
The Aspergillus nidulans alcA promoter drives tightly regulated conditional gene expression in Aspergillus fumigatus permitting validation of essential genes in this human pathogen.Fungal Genet Biol. 2003 Nov;40(2):103-14. doi: 10.1016/s1087-1845(03)00090-2. Fungal Genet Biol. 2003. PMID: 14516763
-
Study of the essentiality of the Aspergillus fumigatus triA gene, encoding RNA triphosphatase, using the heterokaryon rescue technique and the conditional gene expression driven by the alcA and niiA promoters.Fungal Genet Biol. 2010 Jan;47(1):66-79. doi: 10.1016/j.fgb.2009.10.010. Fungal Genet Biol. 2010. PMID: 19883779
-
SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A).Antimicrob Agents Chemother. 2012 Jan;56(1):248-57. doi: 10.1128/AAC.05027-11. Epub 2011 Oct 17. Antimicrob Agents Chemother. 2012. PMID: 22006005 Free PMC article.
-
Genes and molecules involved in Aspergillus fumigatus virulence.Rev Iberoam Micol. 2005 Mar;22(1):1-23. doi: 10.1016/s1130-1406(05)70001-2. Rev Iberoam Micol. 2005. PMID: 15813678 Review.
-
A new experimental murine aspergillosis model to identify strains of Aspergillus fumigatus with reduced virulence.Nihon Ishinkin Gakkai Zasshi. 2002;43(4):203-13. doi: 10.3314/jjmm.43.203. Nihon Ishinkin Gakkai Zasshi. 2002. PMID: 12402022 Review.
Cited by
-
Essential gene discovery in the basidiomycete Cryptococcus neoformans for antifungal drug target prioritization.mBio. 2015 Mar 31;6(2):e02334-14. doi: 10.1128/mBio.02334-14. mBio. 2015. PMID: 25827419 Free PMC article.
-
Ergosterol biosynthesis in Aspergillus fumigatus: its relevance as an antifungal target and role in antifungal drug resistance.Front Microbiol. 2013 Jan 10;3:439. doi: 10.3389/fmicb.2012.00439. eCollection 2012. Front Microbiol. 2013. PMID: 23335918 Free PMC article.
-
OGEE v3: Online GEne Essentiality database with increased coverage of organisms and human cell lines.Nucleic Acids Res. 2021 Jan 8;49(D1):D998-D1003. doi: 10.1093/nar/gkaa884. Nucleic Acids Res. 2021. PMID: 33084874 Free PMC article.
-
Deciphering metabolic traits of the fungal pathogen Aspergillus fumigatus: redundancy vs. essentiality.Front Microbiol. 2012 Dec 3;3:414. doi: 10.3389/fmicb.2012.00414. eCollection 2012. Front Microbiol. 2012. PMID: 23264772 Free PMC article.
-
Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea.Sci Rep. 2018 May 9;8(1):7320. doi: 10.1038/s41598-018-25434-4. Sci Rep. 2018. PMID: 29743510 Free PMC article.
References
-
- Latge JP. The pathobiology of Aspergillus fumigatus . Trends Microbiol. 2001;9:382–389. - PubMed
-
- Tekaia F, Latge JP. Aspergillus fumigatus: Saprophyte or pathogen? Curr Opin Microbiol. 2005;8:385–392. - PubMed
-
- Steinbach WS, Latgé JP, Stevens DA. Advances against aspergillosis. Med Mycol. 2005;43:S1. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

