Control of signaling in a MAP-kinase pathway by an RNA-binding protein
- PMID: 17327913
- PMCID: PMC1803019
- DOI: 10.1371/journal.pone.0000249
Control of signaling in a MAP-kinase pathway by an RNA-binding protein
Abstract
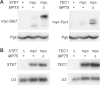
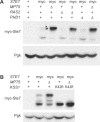
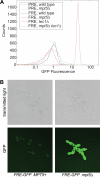
Signaling-protein mRNAs tend to have long untranslated regions (UTRs) containing binding sites for RNA-binding proteins regulating gene expression. Here we show that a PUF-family RNA-binding protein, Mpt5, represses the yeast MAP-kinase pathway controlling differentiation to the filamentous form. Mpt5 represses the protein levels of two pathway components, the Ste7 MAP-kinase kinase and the Tec1 transcriptional activator, and negatively regulates the kinase activity of the Kss1 MAP kinase. Moreover, Mpt5 specifically inhibits the output of the pathway in the absence of stimuli, and thereby prevents inappropriate cell differentiation. The results provide an example of what may be a genome-scale level of regulation at the interface of signaling networks and protein-RNA binding networks.
Conflict of interest statement
Figures



Similar articles
-
Persistent activation by constitutive Ste7 promotes Kss1-mediated invasive growth but fails to support Fus3-dependent mating in yeast.Mol Cell Biol. 2004 Oct;24(20):9221-38. doi: 10.1128/MCB.24.20.9221-9238.2004. Mol Cell Biol. 2004. PMID: 15456892 Free PMC article.
-
Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast.Cell. 2004 Dec 29;119(7):981-90. doi: 10.1016/j.cell.2004.11.053. Cell. 2004. PMID: 15620356
-
Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation.Mol Cell. 2001 Sep;8(3):683-91. doi: 10.1016/s1097-2765(01)00322-7. Mol Cell. 2001. PMID: 11583629 Free PMC article.
-
Fine regulation of Saccharomyces cerevisiae MAPK pathways by post-translational modifications.Yeast. 2010 Aug;27(8):503-11. doi: 10.1002/yea.1791. Yeast. 2010. PMID: 20641029 Review.
-
[Cell surface protein Ecm33 is involved in negative feedback regulation of MAP kinase signalling and development of the in vivo real-time monitoring of MAP kinase signalling].Yakugaku Zasshi. 2011;131(8):1195-200. doi: 10.1248/yakushi.131.1195. Yakugaku Zasshi. 2011. PMID: 21804323 Review. Japanese.
Cited by
-
The yeast PUF protein Puf5 has Pop2-independent roles in response to DNA replication stress.PLoS One. 2010 May 14;5(5):e10651. doi: 10.1371/journal.pone.0010651. PLoS One. 2010. PMID: 20498834 Free PMC article.
-
The RNA-binding protein Whi3 is a key regulator of developmental signaling and ploidy in Saccharomyces cerevisiae.Genetics. 2013 Sep;195(1):73-86. doi: 10.1534/genetics.113.153775. Epub 2013 Jun 14. Genetics. 2013. PMID: 23770701 Free PMC article.
-
Conserved regulation of MAP kinase expression by PUF RNA-binding proteins.PLoS Genet. 2007 Dec 28;3(12):e233. doi: 10.1371/journal.pgen.0030233. PLoS Genet. 2007. PMID: 18166083 Free PMC article.
-
Mutations in the Saccharomyces cerevisiae kinase Cbk1p lead to a fertility defect that can be suppressed by the absence of Brr1p or Mpt5p (Puf5p), proteins involved in RNA metabolism.Genetics. 2009 Sep;183(1):161-73. doi: 10.1534/genetics.109.105130. Epub 2009 Jun 22. Genetics. 2009. PMID: 19546315 Free PMC article.
-
A network of PUF proteins and Ras signaling promote mRNA repression and oogenesis in C. elegans.Dev Biol. 2012 Jun 15;366(2):218-31. doi: 10.1016/j.ydbio.2012.03.019. Epub 2012 Apr 19. Dev Biol. 2012. PMID: 22542599 Free PMC article.
References
-
- Qi M, Elion EA. MAP kinase pathways. J Cell Sci. 2005;118:3569–3572. - PubMed
-
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. - PubMed
-
- Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

