A quaternary SNARE-synaptotagmin-Ca2+-phospholipid complex in neurotransmitter release
- PMID: 17320903
- PMCID: PMC1855161
- DOI: 10.1016/j.jmb.2007.01.040
A quaternary SNARE-synaptotagmin-Ca2+-phospholipid complex in neurotransmitter release
Abstract
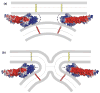
The function of synaptotagmin as a Ca(2+) sensor in neurotransmitter release involves Ca(2+)-dependent phospholipid binding to its two C(2) domains, but this activity alone does not explain why Ca(2+) binding to the C(2)B domain is more critical for release than Ca(2+) binding to the C(2)A domain. Synaptotagmin also binds to SNARE complexes, which are central components of the membrane fusion machinery, and displaces complexins from the SNAREs. However, it is unclear how phospholipid binding to synaptotagmin is coupled to SNARE binding and complexin displacement. Using supported lipid bilayers deposited within microfluidic channels, we now show that Ca(2+) induces simultaneous binding of synaptotagmin to phospholipid membranes and SNARE complexes, resulting in an intimate quaternary complex that we name SSCAP complex. Mutagenesis experiments show that Ca(2+) binding to the C(2)B domain is critical for SSCAP complex formation and displacement of complexin, providing a clear rationale for the preponderant role of the C(2)B domain in release. This and other correlations between the effects of mutations on SSCAP complex formation and their functional effects in vivo suggest a key role for this complex in release. We propose a model whereby the highly positive electrostatic potential at the tip of the SSCAP complex helps to induce membrane fusion during release.
Figures
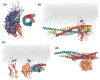
Similar articles
-
A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis.Cell. 2006 Sep 22;126(6):1175-87. doi: 10.1016/j.cell.2006.08.030. Cell. 2006. PMID: 16990140
-
Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution.Nat Struct Mol Biol. 2015 Jul;22(7):555-64. doi: 10.1038/nsmb.3035. Epub 2015 Jun 1. Nat Struct Mol Biol. 2015. PMID: 26030874 Free PMC article.
-
Tomosyn inhibits synaptotagmin-1-mediated step of Ca2+-dependent neurotransmitter release through its N-terminal WD40 repeats.J Biol Chem. 2010 Dec 24;285(52):40943-55. doi: 10.1074/jbc.M110.156893. Epub 2010 Oct 26. J Biol Chem. 2010. PMID: 20978127 Free PMC article.
-
Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release.Trends Cell Biol. 2006 Jul;16(7):339-50. doi: 10.1016/j.tcb.2006.04.006. Epub 2006 May 12. Trends Cell Biol. 2006. PMID: 16698267 Review.
-
Role of SNAREs in membrane fusion.Adv Exp Med Biol. 2011;713:13-32. doi: 10.1007/978-94-007-0763-4_3. Adv Exp Med Biol. 2011. PMID: 21432012 Review.
Cited by
-
Acetylcholinesterase Inhibitors and Drugs Acting on Muscarinic Receptors- Potential Crosstalk of Cholinergic Mechanisms During Pharmacological Treatment.Curr Neuropharmacol. 2017;15(4):637-653. doi: 10.2174/1570159X14666160607212615. Curr Neuropharmacol. 2017. PMID: 27281175 Free PMC article. Review.
-
Synaptotagmin 1 and SNAREs form a complex that is structurally heterogeneous.J Mol Biol. 2011 Jan 21;405(3):696-706. doi: 10.1016/j.jmb.2010.11.015. Epub 2010 Nov 16. J Mol Biol. 2011. PMID: 21087613 Free PMC article.
-
An electrostatically preferred lateral orientation of SNARE complex suggests novel mechanisms for driving membrane fusion.PLoS One. 2010 Jan 26;5(1):e8900. doi: 10.1371/journal.pone.0008900. PLoS One. 2010. PMID: 20126653 Free PMC article.
-
A fast, single-vesicle fusion assay mimics physiological SNARE requirements.Proc Natl Acad Sci U S A. 2010 Feb 23;107(8):3517-21. doi: 10.1073/pnas.0914723107. Epub 2010 Feb 2. Proc Natl Acad Sci U S A. 2010. PMID: 20133592 Free PMC article.
-
Titration of Syntaxin1 in mammalian synapses reveals multiple roles in vesicle docking, priming, and release probability.J Neurosci. 2013 Oct 16;33(42):16698-714. doi: 10.1523/JNEUROSCI.0187-13.2013. J Neurosci. 2013. PMID: 24133272 Free PMC article.
References
-
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. - PubMed
-
- Fernandez-Chacon R, Konigstorfer A, Gerber SH, Garcia J, Matos MF, Stevens CF, Brose N, Rizo J, Rosenmund C, Sudhof TC. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. - PubMed
-
- Mackler JM, Drummond JA, Loewen CA, Robinson IM, Reist NE. The C(2)B Ca(2+)-binding motif of synaptotagmin is required for synaptic transmission in vivo. Nature. 2002;418:340–344. - PubMed
-
- Bai J, Chapman ER. The C2 domains of synaptotagmin--partners in exocytosis. Trends Biochem Sci. 2004;29:143–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

