Microtubule disruption stimulates P-body formation
- PMID: 17307817
- PMCID: PMC1831866
- DOI: 10.1261/rna.355807
Microtubule disruption stimulates P-body formation
Abstract
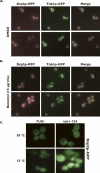
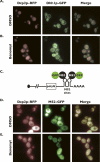
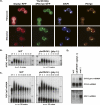
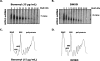
Processing bodies (P-bodies) are subcellular ribonucleoprotein (RNP) granules that have been hypothesized to be sites of mRNA degradation, mRNA translational control, and/or mRNA storage. Importantly, P-bodies are conserved from yeast to mammals and contain a common set of evolutionarily conserved protein constituents. P-bodies are dynamic structures and their formation appears to fluctuate in correlation with alterations in mRNA metabolism. Despite these observations, little is understood about how P-body structures are formed within the cell. In this study, we demonstrate a relationship between P-bodies and microtubules in the budding yeast, Saccharomyces cerevisiae. First, we demonstrate that disruption of microtubules by treatment with the drug benomyl leads to aggregation of P-body components. Consistent with this finding, we also demonstrate that disruption of microtubules by a temperature-sensitive allele of the major alpha tubulin, TUB1 (tub1-724) stimulates P-body formation. Second, we find that the alpha-tubulin protein Tub1 colocalizes with P-bodies upon microtubule destabilization. Third, we determine that a putative tubulin tyrosine ligase, encoded by YBR094W, is a protein component of P-bodies, providing additional evidence for a physical connection between P-bodies and microtubules. Finally, we establish that P-bodies formed by microtubule destabilization fail to correlate with global changes in the stability of mRNA or in general mRNA translation. These findings demonstrate that the aggregation of P-body components is linked to the intracellular microtubule network, and, further, that P-bodies formed by disruption of microtubules aggregate independent of broad alterations in either mRNA decay or mRNA translation.
Figures
Similar articles
-
Microtubule stability in budding yeast: characterization and dosage suppression of a benomyl-dependent tubulin mutant.Mol Biol Cell. 1995 Sep;6(9):1241-59. doi: 10.1091/mbc.6.9.1241. Mol Biol Cell. 1995. PMID: 8534919 Free PMC article.
-
Single site alpha-tubulin mutation affects astral microtubules and nuclear positioning during anaphase in Saccharomyces cerevisiae: possible role for palmitoylation of alpha-tubulin.Mol Biol Cell. 2001 Sep;12(9):2672-87. doi: 10.1091/mbc.12.9.2672. Mol Biol Cell. 2001. PMID: 11553707 Free PMC article.
-
A novel protein complex promoting formation of functional alpha- and gamma-tubulin.EMBO J. 1998 Feb 16;17(4):952-66. doi: 10.1093/emboj/17.4.952. EMBO J. 1998. PMID: 9463374 Free PMC article.
-
Regulating microtubule properties by modifying their organizing minus ends.Mol Cell. 2001 Nov;8(5):931-2. doi: 10.1016/s1097-2765(01)00394-x. Mol Cell. 2001. PMID: 11741527 Review.
-
Composition of the spindle pole body of Saccharomyces cerevisiae and the proteins involved in its duplication.Curr Genet. 2002 Feb;40(5):291-310. doi: 10.1007/s00294-001-0263-x. Epub 2001 Dec 8. Curr Genet. 2002. PMID: 11935220 Review.
Cited by
-
Biogenesis, turnover, and mode of action of plant microRNAs.Plant Cell. 2013 Jul;25(7):2383-99. doi: 10.1105/tpc.113.113159. Epub 2013 Jul 23. Plant Cell. 2013. PMID: 23881412 Free PMC article. Review.
-
RNAi Screen Identifies Novel Regulators of RNP Granules in the Caenorhabditis elegans Germ Line.G3 (Bethesda). 2016 Aug 9;6(8):2643-54. doi: 10.1534/g3.116.031559. G3 (Bethesda). 2016. PMID: 27317775 Free PMC article.
-
Myosin Va is required for P body but not stress granule formation.J Biol Chem. 2011 Apr 1;286(13):11519-28. doi: 10.1074/jbc.M110.182808. Epub 2011 Jan 18. J Biol Chem. 2011. PMID: 21245139 Free PMC article.
-
Mitochondria associate with P-bodies and modulate microRNA-mediated RNA interference.J Biol Chem. 2011 Jul 8;286(27):24219-30. doi: 10.1074/jbc.M111.240259. Epub 2011 May 16. J Biol Chem. 2011. PMID: 21576251 Free PMC article.
-
Somatic insulin signaling regulates a germline starvation response in Drosophila egg chambers.Dev Biol. 2015 Feb 15;398(2):206-17. doi: 10.1016/j.ydbio.2014.11.021. Epub 2014 Dec 3. Dev Biol. 2015. PMID: 25481758 Free PMC article.
References
-
- Bhattacharyya, S.N., Habermacher, R., Martine, U., Closs, E.I., Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
