The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement
- PMID: 17296725
- PMCID: PMC1899931
- DOI: 10.1128/MCB.02190-06
The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement
Abstract
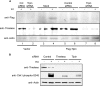
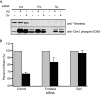
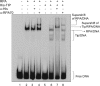
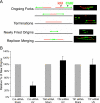
UV-induced DNA damage stalls DNA replication forks and activates the intra-S checkpoint to inhibit replicon initiation. In response to stalled replication forks, ATR phosphorylates and activates the transducer kinase Chk1 through interactions with the mediator proteins TopBP1, Claspin, and Timeless (Tim). Murine Tim recently was shown to form a complex with Tim-interacting protein (Tipin), and a similar complex was shown to exist in human cells. Knockdown of Tipin using small interfering RNA reduced the expression of Tim and reversed the intra-S checkpoint response to UVC. Tipin interacted with replication protein A (RPA) and RPA-coated DNA, and RPA promoted the loading of Tipin onto RPA-free DNA. Immunofluorescence analysis of spread DNA fibers showed that treating HeLa cells with 2.5 J/m(2) UVC not only inhibited the initiation of new replicons but also reduced the rate of chain elongation at active replication forks. The depletion of Tim and Tipin reversed the UV-induced inhibition of replicon initiation but affected the rate of DNA synthesis at replication forks in different ways. In undamaged cells depleted of Tim, the apparent rate of replication fork progression was 52% of the control. In contrast, Tipin depletion had little or no effect on fork progression in unirradiated cells but significantly attenuated the UV-induced inhibition of DNA chain elongation. Together, these findings indicate that the Tim-Tipin complex mediates the UV-induced intra-S checkpoint, Tim is needed to maintain DNA replication fork movement in the absence of damage, Tipin interacts with RPA on DNA and, in UV-damaged cells, Tipin slows DNA chain elongation in active replicons.
Figures


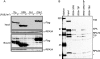
Similar articles
-
Timeless functions independently of the Tim-Tipin complex to promote sister chromatid cohesion in normal human fibroblasts.Cell Cycle. 2011 May 15;10(10):1618-24. doi: 10.4161/cc.10.10.15613. Epub 2011 May 15. Cell Cycle. 2011. PMID: 21508667 Free PMC article.
-
Tim-Tipin dysfunction creates an indispensible reliance on the ATR-Chk1 pathway for continued DNA synthesis.J Cell Biol. 2009 Oct 5;187(1):15-23. doi: 10.1083/jcb.200905006. J Cell Biol. 2009. PMID: 19805627 Free PMC article.
-
Separation of intra-S checkpoint protein contributions to DNA replication fork protection and genomic stability in normal human fibroblasts.Cell Cycle. 2013 Jan 15;12(2):332-45. doi: 10.4161/cc.23177. Epub 2012 Jan 15. Cell Cycle. 2013. PMID: 23255133 Free PMC article.
-
TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation.Cell Cycle. 2009 Sep 15;8(18):2877-84. doi: 10.4161/cc.8.18.9485. Epub 2009 Sep 9. Cell Cycle. 2009. PMID: 19652550 Review.
-
The human intra-S checkpoint response to UVC-induced DNA damage.Carcinogenesis. 2010 May;31(5):751-65. doi: 10.1093/carcin/bgp230. Epub 2009 Sep 30. Carcinogenesis. 2010. PMID: 19793801 Free PMC article. Review.
Cited by
-
Activation of the ATR kinase by the RPA-binding protein ETAA1.Nat Cell Biol. 2016 Nov;18(11):1196-1207. doi: 10.1038/ncb3422. Epub 2016 Oct 10. Nat Cell Biol. 2016. PMID: 27723717
-
RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe.Mol Biol Cell. 2008 Feb;19(2):595-607. doi: 10.1091/mbc.e07-06-0618. Epub 2007 Nov 28. Mol Biol Cell. 2008. PMID: 18045993 Free PMC article.
-
Multiple ATR-Chk1 pathway proteins preferentially associate with checkpoint-inducing DNA substrates.PLoS One. 2011;6(7):e22986. doi: 10.1371/journal.pone.0022986. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829571 Free PMC article.
-
Recombinase and translesion DNA polymerase decrease the speed of replication fork progression during the DNA damage response in Escherichia coli cells.Nucleic Acids Res. 2015 Feb 18;43(3):1714-25. doi: 10.1093/nar/gkv044. Epub 2015 Jan 27. Nucleic Acids Res. 2015. PMID: 25628359 Free PMC article.
-
The Fork Protection Complex: A Regulatory Hub at the Head of the Replisome.Subcell Biochem. 2022;99:83-107. doi: 10.1007/978-3-031-00793-4_3. Subcell Biochem. 2022. PMID: 36151374
References
-
- Aten, J. A., P. J. Bakker, J. Stap, G. A. Boschman, and C. H. Veenhof. 1992. DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem. J. 24:251-259. - PubMed
-
- Bassett, E., N. M. King, M. F. Bryant, S. Hector, L. Pendyala, S. G. Chaney, and M. Cordeiro-Stone. 2004. The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res 64:6469-6475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
