Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast
- PMID: 17261596
- PMCID: PMC1899950
- DOI: 10.1128/MCB.01039-06
Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast
Abstract
Fission yeast has two TOR (target of rapamycin) kinases, namely Tor1 and Tor2. Tor1 is required for survival under stressed conditions, proper G(1) arrest, and sexual development. In contrast, Tor2 is essential for growth. To analyze the functions of Tor2, we constructed two temperature-sensitive tor2 mutants. Interestingly, at the restrictive temperature, these mutants mimicked nitrogen starvation by arresting the cell cycle in G(1) phase and initiating sexual development. Microarray analysis indicated that expression of nitrogen starvation-responsive genes was induced extensively when Tor2 function was suppressed, suggesting that Tor2 normally mediates a signal from the nitrogen source. As with mammalian and budding yeast TOR, we find that fission yeast TOR also forms multiprotein complexes analogous to TORC1 and TORC2. The raptor homologue, Mip1, likely forms a complex predominantly with Tor2, producing TORC1. The rictor/Avo3 homologue, Ste20, and the Avo1 homologue, Sin1, appear to form TORC2 mainly with Tor1 but may also bind Tor2. The Lst8 homologue, Wat1, binds to both Tor1 and Tor2. Our analysis shows, with respect to promotion of G(1) arrest and sexual development, that the loss of Tor1 (TORC2) and the loss of Tor2 (TORC1) exhibit opposite effects. This highlights an intriguing functional relationship among TOR kinase complexes in the fission yeast Schizosaccharomyces pombe.
Figures
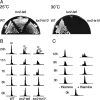
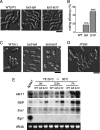

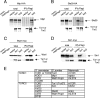
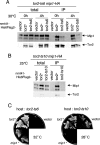
Similar articles
-
Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast.Genetics. 2007 Mar;175(3):1153-62. doi: 10.1534/genetics.106.064170. Epub 2006 Dec 18. Genetics. 2007. PMID: 17179073 Free PMC article.
-
Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits.Genes Cells. 2007 Dec;12(12):1357-70. doi: 10.1111/j.1365-2443.2007.01141.x. Genes Cells. 2007. PMID: 18076573
-
Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase.Genes Cells. 2006 Dec;11(12):1367-79. doi: 10.1111/j.1365-2443.2006.01025.x. Genes Cells. 2006. PMID: 17121544
-
TOR signaling in fission yeast.Crit Rev Biochem Mol Biol. 2008 Jul-Aug;43(4):277-83. doi: 10.1080/10409230802254911. Crit Rev Biochem Mol Biol. 2008. PMID: 18756382 Review.
-
TORC1-Dependent Phosphorylation Targets in Fission Yeast.Biomolecules. 2017 Jul 3;7(3):50. doi: 10.3390/biom7030050. Biomolecules. 2017. PMID: 28671615 Free PMC article. Review.
Cited by
-
TOR complex 2 localises to the cytokinetic actomyosin ring and controls the fidelity of cytokinesis.J Cell Sci. 2016 Jul 1;129(13):2613-24. doi: 10.1242/jcs.190124. Epub 2016 May 20. J Cell Sci. 2016. PMID: 27206859 Free PMC article.
-
Rab-family GTPase regulates TOR complex 2 signaling in fission yeast.Curr Biol. 2010 Nov 23;20(22):1975-82. doi: 10.1016/j.cub.2010.10.026. Epub 2010 Oct 28. Curr Biol. 2010. PMID: 21035342 Free PMC article.
-
The GATA Transcription Factor Gaf1 Represses tRNAs, Inhibits Growth, and Extends Chronological Lifespan Downstream of Fission Yeast TORC1.Cell Rep. 2020 Mar 10;30(10):3240-3249.e4. doi: 10.1016/j.celrep.2020.02.058. Cell Rep. 2020. PMID: 32160533 Free PMC article.
-
Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control.Cell Cycle. 2008 Feb 1;7(3):358-64. doi: 10.4161/cc.7.3.5245. Epub 2007 Nov 1. Cell Cycle. 2008. PMID: 18235227 Free PMC article.
-
Glucose activates TORC2-Gad8 protein via positive regulation of the cAMP/cAMP-dependent protein kinase A (PKA) pathway and negative regulation of the Pmk1 protein-mitogen-activated protein kinase pathway.J Biol Chem. 2014 Aug 1;289(31):21727-37. doi: 10.1074/jbc.M114.573824. Epub 2014 Jun 13. J Biol Chem. 2014. PMID: 24928510 Free PMC article.
References
-
- Abraham, R. T. 2004. PI 3-kinase related kinases: “big” players in stress-induced signaling pathways. DNA Repair 3:883-887. - PubMed
-
- Álvarez, B., and S. J. Moreno. 2006. Fission yeast Tor2 promotes cell growth and represses cell differentiation. Cell Sci. 119:4475-4485. - PubMed
-
- Aspuria, P. J., and F. Tamanoi. 2004. The Rheb family of GTP-binding proteins. Cell Signal 16:1105-1112. - PubMed
-
- Averbeck, N., S. Sunder, N. Sample, J. A. Wise, and J. Leatherwood. 2005. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol. Cell 18:491-498. - PubMed
-
- Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
