Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations
- PMID: 17251351
- PMCID: PMC1785253
- DOI: 10.1073/pnas.0607097104
Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations
Abstract
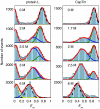
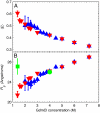
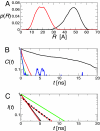
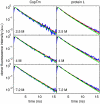
To obtain quantitative information on the size and dynamics of unfolded proteins we combined single-molecule lifetime and intensity FRET measurements with molecular simulations. We compared the unfolded states of the 64-residue, alpha/beta protein L and the 66-residue, all-beta cold-shock protein CspTm. The average radius of gyration (Rg) calculated from FRET data on freely diffusing molecules was identical for the two unfolded proteins at guanidinium chloride concentrations >3 M, and the FRET-derived Rg of protein L agreed well with the Rg previously measured by equilibrium small-angle x-ray scattering. As the denaturant concentration was lowered, the mean FRET efficiency of the unfolded subpopulation increased, signaling collapse of the polypeptide chain, with protein L being slightly more compact than CspTm. A decrease in Rg with decreasing denaturant was also observed in all-atom molecular dynamics calculations in explicit water/urea solvent, and Langevin simulations of a simplified representation of the polypeptide suggest that collapse can result from either increased interresidue attraction or decreased excluded volume. In contrast to both the FRET and simulation results, previous time-resolved small-angle x-ray scattering experiments showed no collapse for protein L. Analysis of the donor fluorescence decay of the unfolded subpopulation of both proteins gives information about the end-to-end chain distribution and suggests that chain dynamics is slow compared with the donor life-time of approximately 2 ns, whereas the bin-size independence of the small excess width above the shot noise for the FRET efficiency distributions may result from incomplete conformational averaging on even the 1-ms time scale.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Folding of Protein L with Implications for Collapse in the Denatured State Ensemble.J Am Chem Soc. 2016 Mar 2;138(8):2609-16. doi: 10.1021/jacs.5b11300. Epub 2016 Feb 16. J Am Chem Soc. 2016. PMID: 26835789
-
Single-molecule FRET study of denaturant induced unfolding of RNase H.J Mol Biol. 2006 Mar 17;357(1):313-24. doi: 10.1016/j.jmb.2005.12.061. Epub 2006 Jan 9. J Mol Biol. 2006. PMID: 16426636
-
Small-angle X-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state.J Mol Biol. 2012 May 4;418(3-4):226-36. doi: 10.1016/j.jmb.2012.01.016. Epub 2012 Jan 27. J Mol Biol. 2012. PMID: 22306460 Free PMC article.
-
Atomic-level characterization of disordered protein ensembles.Curr Opin Struct Biol. 2007 Feb;17(1):3-14. doi: 10.1016/j.sbi.2007.01.009. Epub 2007 Jan 23. Curr Opin Struct Biol. 2007. PMID: 17250999 Review.
-
The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements.Chemphyschem. 2005 May;6(5):858-70. doi: 10.1002/cphc.200400617. Chemphyschem. 2005. PMID: 15884068 Review.
Cited by
-
Conformational properties of the unfolded state of Im7 in nondenaturing conditions.J Mol Biol. 2012 Feb 17;416(2):300-18. doi: 10.1016/j.jmb.2011.12.041. Epub 2011 Dec 28. J Mol Biol. 2012. PMID: 22226836 Free PMC article.
-
Multiple conformations of full-length p53 detected with single-molecule fluorescence resonance energy transfer.Proc Natl Acad Sci U S A. 2009 Dec 8;106(49):20758-63. doi: 10.1073/pnas.0909644106. Epub 2009 Nov 20. Proc Natl Acad Sci U S A. 2009. PMID: 19933326 Free PMC article.
-
Generating Ensembles of Dynamic Misfolding Proteins.Front Neurosci. 2022 Mar 31;16:881534. doi: 10.3389/fnins.2022.881534. eCollection 2022. Front Neurosci. 2022. PMID: 35431773 Free PMC article. Review.
-
Single molecule characterization of α-synuclein in aggregation-prone states.Biophys J. 2010 Nov 3;99(9):3048-55. doi: 10.1016/j.bpj.2010.08.056. Biophys J. 2010. PMID: 21044603 Free PMC article.
-
Probing the mechanism of inhibition of amyloid-β(1-42)-induced neurotoxicity by the chaperonin GroEL.Proc Natl Acad Sci U S A. 2018 Dec 18;115(51):E11924-E11932. doi: 10.1073/pnas.1817477115. Epub 2018 Dec 3. Proc Natl Acad Sci U S A. 2018. PMID: 30509980 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

