RNA viruses: hijacking the dynamic nucleolus
- PMID: 17224921
- PMCID: PMC7097444
- DOI: 10.1038/nrmicro1597
RNA viruses: hijacking the dynamic nucleolus
Abstract
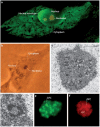
The nucleolus is a dynamic subnuclear structure with roles in ribosome subunit biogenesis, mediation of cell-stress responses and regulation of cell growth. The proteome and structure of the nucleolus are constantly changing in response to metabolic conditions. RNA viruses interact with the nucleolus to usurp host-cell functions and recruit nucleolar proteins to facilitate virus replication. Investigating the interactions between RNA viruses and the nucleolus will facilitate the design of novel anti-viral therapies, such as recombinant vaccines and therapeutic molecular interventions, and also contribute to a more detailed understanding of the cell biology of the nucleolus.
Conflict of interest statement
The author declares no competing financial interests.
Figures
Similar articles
-
Involvement of the plant nucleolus in virus and viroid infections: parallels with animal pathosystems.Adv Virus Res. 2010;77:119-58. doi: 10.1016/B978-0-12-385034-8.00005-3. Adv Virus Res. 2010. PMID: 20951872 Free PMC article. Review.
-
The nucleolar interface of RNA viruses.Cell Microbiol. 2015 Aug;17(8):1108-20. doi: 10.1111/cmi.12465. Epub 2015 Jun 26. Cell Microbiol. 2015. PMID: 26041433 Review.
-
Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus.Cell Microbiol. 2006 Jul;8(7):1147-57. doi: 10.1111/j.1462-5822.2006.00698.x. Cell Microbiol. 2006. PMID: 16819967 Free PMC article.
-
The nucleolus--a gateway to viral infection?Arch Virol. 2002 Jun;147(6):1077-89. doi: 10.1007/s00705-001-0792-0. Arch Virol. 2002. PMID: 12111420 Free PMC article. Review.
-
Viruses and the nucleolus: the fatal attraction.Biochim Biophys Acta. 2014 Jun;1842(6):840-7. doi: 10.1016/j.bbadis.2013.12.010. Epub 2013 Dec 27. Biochim Biophys Acta. 2014. PMID: 24378568 Free PMC article.
Cited by
-
Proteomics analysis of the nucleolus in adenovirus-infected cells.Mol Cell Proteomics. 2010 Jan;9(1):117-30. doi: 10.1074/mcp.M900338-MCP200. Epub 2009 Oct 7. Mol Cell Proteomics. 2010. PMID: 19812395 Free PMC article.
-
Adeno-associated virus type 2 (AAV2) uncoating is a stepwise process and is linked to structural reorganization of the nucleolus.PLoS Pathog. 2022 Jul 11;18(7):e1010187. doi: 10.1371/journal.ppat.1010187. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35816507 Free PMC article.
-
Chromatin: linking structure and function in the nucleolus.Chromosoma. 2009 Feb;118(1):11-23. doi: 10.1007/s00412-008-0184-2. Epub 2008 Oct 17. Chromosoma. 2009. PMID: 18925405 Review.
-
Nucleolin mediates SARS-CoV-2 replication and viral-induced apoptosis of host cells.Antiviral Res. 2023 Mar;211:105550. doi: 10.1016/j.antiviral.2023.105550. Epub 2023 Feb 3. Antiviral Res. 2023. PMID: 36740097 Free PMC article.
-
The bovine immunodeficiency virus rev protein: identification of a novel lentiviral bipartite nuclear localization signal harboring an atypical spacer sequence.J Virol. 2009 Dec;83(24):12842-53. doi: 10.1128/JVI.01613-09. Epub 2009 Oct 14. J Virol. 2009. PMID: 19828621 Free PMC article.
References
-
- Hernandez-Verdun D, Roussel P, Gebrane-Younes J. Emerging concepts of nucleolar assembly. J. Cell Sci. 2002;115:2265–2270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

