UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment
- PMID: 17215291
- PMCID: PMC1866064
- DOI: 10.1128/JVI.02201-06
UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment
Abstract
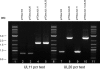
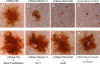
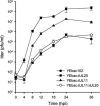
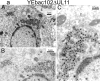
Egress of herpes simplex virus type 1 (HSV-1) from the nucleus of the infected cell to extracellular spaces involves a number of distinct steps, including primary envelopment by budding into the perinuclear space, de-envelopment into the cytoplasm, cytoplasmic reenvelopment, and translocation of enveloped virions to extracellular spaces. UL20/gK-null viruses are blocked in cytoplasmic virion envelopment and egress, as indicated by an accumulation of unenveloped or partially enveloped capsids in the cytoplasm. Similarly, UL11-null mutants accumulate unenveloped capsids in the cytoplasm. To assess whether UL11 and UL20/gK function independently or synergistically in cytoplasmic envelopment, recombinant viruses having either the UL20 or UL11 gene deleted were generated. In addition, a recombinant virus containing a deletion of both UL20 and UL11 genes was constructed using the HSV-1(F) genome cloned into a bacterial artificial chromosome. Ultrastructural examination of virus-infected cells showed that both UL20- and UL11-null viruses accumulated unenveloped capsids in the cytoplasm. However, the morphology and distribution of the accumulated capsids appeared to be distinct, with the UL11-null virions forming aggregates of capsids having diffuse tegument-derived material and the UL20-null virus producing individual capsids in close juxtaposition to cytoplasmic membranes. The UL20/UL11 double-null virions appeared morphologically similar to the UL20-null viruses. Experiments on the kinetics of viral replication revealed that the UL20/UL11 double-null virus replicated in a manner similar to the UL20-null virus. Additional experiments revealed that transiently expressed UL11 localized to the trans-Golgi network (TGN) independently of either gK or UL20. Furthermore, virus infection with the UL11/UL20 double-null virus did not alter the TGN localization of transiently expressed UL11 or UL20 proteins, indicating that these proteins did not interact. Taken together, these results show that the intracellular transport and TGN localization of UL11 is independent of UL20/gK functions, and that UL20/gK are required and function prior to UL11 protein in virion cytoplasmic envelopment.
Figures



Similar articles
-
Herpes simplex virus 1 protein UL37 interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment.J Virol. 2014 Jun;88(11):5927-35. doi: 10.1128/JVI.00278-14. Epub 2014 Mar 5. J Virol. 2014. PMID: 24600000 Free PMC article.
-
Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment.J Virol. 2005 Jan;79(1):299-313. doi: 10.1128/JVI.79.1.299-313.2005. J Virol. 2005. PMID: 15596825 Free PMC article.
-
Functional hierarchy of herpes simplex virus 1 viral glycoproteins in cytoplasmic virion envelopment and egress.J Virol. 2012 Apr;86(8):4262-70. doi: 10.1128/JVI.06766-11. Epub 2012 Feb 8. J Virol. 2012. PMID: 22318149 Free PMC article.
-
Herpesvirus Nuclear Egress across the Outer Nuclear Membrane.Viruses. 2021 Nov 24;13(12):2356. doi: 10.3390/v13122356. Viruses. 2021. PMID: 34960625 Free PMC article. Review.
-
Tegument Assembly and Secondary Envelopment of Alphaherpesviruses.Viruses. 2015 Sep 18;7(9):5084-114. doi: 10.3390/v7092861. Viruses. 2015. PMID: 26393641 Free PMC article. Review.
Cited by
-
Egress of HSV-1 capsid requires the interaction of VP26 and a cellular tetraspanin membrane protein.Virol J. 2010 Jul 14;7:156. doi: 10.1186/1743-422X-7-156. Virol J. 2010. PMID: 20630051 Free PMC article.
-
Interaction and interdependent packaging of tegument protein UL11 and glycoprotein e of herpes simplex virus.J Virol. 2011 Sep;85(18):9437-46. doi: 10.1128/JVI.05207-11. Epub 2011 Jul 6. J Virol. 2011. PMID: 21734040 Free PMC article.
-
The herpes simplex virus type 1 glycoprotein D (gD) cytoplasmic terminus and full-length gE are not essential and do not function in a redundant manner for cytoplasmic virion envelopment and egress.J Virol. 2009 Jun;83(12):6115-24. doi: 10.1128/JVI.00128-09. Epub 2009 Apr 8. J Virol. 2009. PMID: 19357164 Free PMC article.
-
Myristylation and palmitylation of HSV-1 UL11 are not essential for its function.Virology. 2010 Feb 5;397(1):80-8. doi: 10.1016/j.virol.2009.10.046. Epub 2009 Nov 26. Virology. 2010. PMID: 19944438 Free PMC article.
-
Herpes simplex virus 1 protein UL37 interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment.J Virol. 2014 Jun;88(11):5927-35. doi: 10.1128/JVI.00278-14. Epub 2014 Mar 5. J Virol. 2014. PMID: 24600000 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

