The intracellular sites of early replication and budding of SARS-coronavirus
- PMID: 17210170
- PMCID: PMC7103305
- DOI: 10.1016/j.virol.2006.11.027
The intracellular sites of early replication and budding of SARS-coronavirus
Abstract
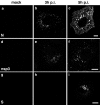
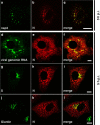
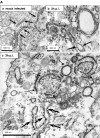
In this study, we analyzed the replication and budding sites of severe acute respiratory syndrome coronavirus (SARS-CoV) at early time points of infection. We detected cytoplasmic accumulations containing the viral nucleocapsid protein, viral RNA and the non-structural protein nsp3. Using EM techniques, we found that these putative viral replication sites were associated with characteristic membrane tubules and double membrane vesicles that most probably originated from ER cisternae. In addition to its presence at the replication sites, N also accumulated in the Golgi region and colocalized with the viral spike protein. Immuno-EM revealed that budding occurred at membranes of the ERGIC (ER-Golgi intermediate compartment) and the Golgi region as early as 3 h post infection, demonstrating that SARS-CoV replicates surprisingly fast. Our data suggest that SARS-CoV establishes replication complexes at ER-derived membranes. Later on, viral nucleocapsids have to be transported to the budding sites in the Golgi region where the viral glycoproteins accumulate and particle formation occurs.
Figures



Similar articles
-
The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein.J Virol. 2007 Mar;81(5):2418-28. doi: 10.1128/JVI.02146-06. Epub 2006 Dec 13. J Virol. 2007. PMID: 17166901 Free PMC article.
-
Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication.J Virol. 2007 Nov;81(22):12323-36. doi: 10.1128/JVI.01506-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855519 Free PMC article.
-
Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication.mBio. 2017 Nov 21;8(6):e01658-17. doi: 10.1128/mBio.01658-17. mBio. 2017. PMID: 29162711 Free PMC article.
-
Hosting the severe acute respiratory syndrome coronavirus: specific cell factors required for infection.Cell Microbiol. 2006 Aug;8(8):1211-8. doi: 10.1111/j.1462-5822.2006.00744.x. Epub 2006 Jun 27. Cell Microbiol. 2006. PMID: 16803585 Free PMC article. Review.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
Cited by
-
Calcium‐dependent antimicrobials: Nature‐inspired materials and designs.Exploration (Beijing). 2024 Mar 12;4(5):20230099. doi: 10.1002/EXP.20230099. eCollection 2024 Oct. Exploration (Beijing). 2024. PMID: 39439493 Free PMC article. Review. Catalan.
-
Coronavirus envelope protein activates TMED10-mediated unconventional secretion of inflammatory factors.Nat Commun. 2024 Oct 8;15(1):8708. doi: 10.1038/s41467-024-52818-0. Nat Commun. 2024. PMID: 39379362 Free PMC article.
-
N terminus of SARS-CoV-2 nonstructural protein 3 interrupts RNA-driven phase separation of N protein by displacing RNA.J Biol Chem. 2024 Nov;300(11):107828. doi: 10.1016/j.jbc.2024.107828. Epub 2024 Sep 26. J Biol Chem. 2024. PMID: 39341499 Free PMC article.
-
AlphaFold2 Reveals Structural Patterns of Seasonal Haplotype Diversification in SARS-CoV-2 Nucleocapsid Protein Variants.Viruses. 2024 Aug 25;16(9):1358. doi: 10.3390/v16091358. Viruses. 2024. PMID: 39339835 Free PMC article.
-
A specific phosphorylation-dependent conformational switch in SARS-CoV-2 nucleocapsid protein inhibits RNA binding.Sci Adv. 2024 Aug 2;10(31):eaax2323. doi: 10.1126/sciadv.aax2323. Epub 2024 Aug 2. Sci Adv. 2024. PMID: 39093972 Free PMC article.
References
-
- Blasco R., Moss B. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene. 1995;158(2):157–162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

