Powerful multilocus tests of genetic association in the presence of gene-gene and gene-environment interactions
- PMID: 17186459
- PMCID: PMC1698705
- DOI: 10.1086/509704
Powerful multilocus tests of genetic association in the presence of gene-gene and gene-environment interactions
Abstract
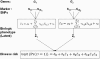
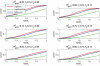
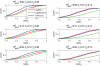
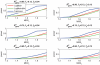
In modern genetic epidemiology studies, the association between the disease and a genomic region, such as a candidate gene, is often investigated using multiple SNPs. We propose a multilocus test of genetic association that can account for genetic effects that might be modified by variants in other genes or by environmental factors. We consider use of the venerable and parsimonious Tukey's 1-degree-of-freedom model of interaction, which is natural when individual SNPs within a gene are associated with disease through a common biological mechanism; in contrast, many standard regression models are designed as if each SNP has unique functional significance. On the basis of Tukey's model, we propose a novel but computationally simple generalized test of association that can simultaneously capture both the main effects of the variants within a genomic region and their interactions with the variants in another region or with an environmental exposure. We compared performance of our method with that of two standard tests of association, one ignoring gene-gene/gene-environment interactions and the other based on a saturated model of interactions. We demonstrate major power advantages of our method both in analysis of data from a case-control study of the association between colorectal adenoma and DNA variants in the NAT2 genomic region, which are well known to be related to a common biological phenotype, and under different models of gene-gene interactions with use of simulated data.
Figures


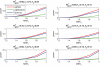

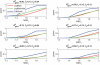
Similar articles
-
Retrospective analysis of haplotype-based case control studies under a flexible model for gene environment association.Biostatistics. 2008 Jan;9(1):81-99. doi: 10.1093/biostatistics/kxm011. Epub 2007 May 8. Biostatistics. 2008. PMID: 17490987 Free PMC article.
-
Characterization of N-acetyltransferase 1 and 2 polymorphisms and haplotype analysis for inflammatory bowel disease and sporadic colorectal carcinoma.BMC Med Genet. 2007 May 30;8:28. doi: 10.1186/1471-2350-8-28. BMC Med Genet. 2007. PMID: 17537267 Free PMC article.
-
Effect of SULT1A1 and NAT2 genetic polymorphism on the association between cigarette smoking and colorectal adenomas.Int J Cancer. 2004 Jan 1;108(1):97-103. doi: 10.1002/ijc.11533. Int J Cancer. 2004. PMID: 14618622
-
N-acetyltransferase polymorphisms and colorectal cancer: a HuGE review.Am J Epidemiol. 2000 May 1;151(9):846-61. doi: 10.1093/oxfordjournals.aje.a010289. Am J Epidemiol. 2000. PMID: 10791558 Review.
-
A meta-analysis of the association of N-acetyltransferase 2 gene (NAT2) variants with breast cancer.Am J Epidemiol. 2007 Aug 1;166(3):246-54. doi: 10.1093/aje/kwm066. Epub 2007 May 29. Am J Epidemiol. 2007. PMID: 17535831 Review.
Cited by
-
Adaptive tests for detecting gene-gene and gene-environment interactions.Hum Hered. 2011;72(2):98-109. doi: 10.1159/000330632. Epub 2011 Sep 16. Hum Hered. 2011. PMID: 21934325 Free PMC article.
-
Challenges and opportunities in genome-wide environmental interaction (GWEI) studies.Hum Genet. 2012 Oct;131(10):1591-613. doi: 10.1007/s00439-012-1192-0. Epub 2012 Jul 4. Hum Genet. 2012. PMID: 22760307 Free PMC article. Review.
-
A random forest approach to the detection of epistatic interactions in case-control studies.BMC Bioinformatics. 2009 Jan 30;10 Suppl 1(Suppl 1):S65. doi: 10.1186/1471-2105-10-S1-S65. BMC Bioinformatics. 2009. PMID: 19208169 Free PMC article.
-
Mutual information for testing gene-environment interaction.PLoS One. 2009;4(2):e4578. doi: 10.1371/journal.pone.0004578. Epub 2009 Feb 24. PLoS One. 2009. PMID: 19238204 Free PMC article.
-
Epistatic module detection for case-control studies: a Bayesian model with a Gibbs sampling strategy.PLoS Genet. 2009 May;5(5):e1000464. doi: 10.1371/journal.pgen.1000464. Epub 2009 May 1. PLoS Genet. 2009. PMID: 19412524 Free PMC article.
References
Web Resources
-
- NCI Advanced Technology Center, http://cgf.nci.nih.gov/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NAT2, GPX3, GPX4, and colorectal cancer)
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

