RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin
- PMID: 17178837
- PMCID: PMC1820475
- DOI: 10.1128/MCB.01956-06
RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin
Abstract
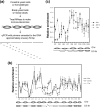
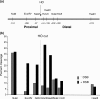
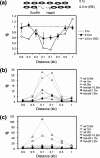
Repair of DNA double-strand breaks (DSBs) protects cells and organisms, as well as their genome integrity. Since DSB repair occurs in the context of chromatin, chromatin must be modified to prevent it from inhibiting DSB repair. Evidence supports the role of histone modifications and ATP-dependent chromatin remodeling in repair and signaling of chromosome DSBs. The key questions are, then, what the nature of chromatin altered by DSBs is and how remodeling of chromatin facilitates DSB repair. Here we report a chromatin alteration caused by a single HO endonuclease-generated DSB at the Saccharomyces cerevisiae MAT locus. The break induces rapid nucleosome migration to form histone-free DNA of a few hundred base pairs immediately adjacent to the break. The DSB-induced nucleosome repositioning appears independent of end processing, since it still occurs when the 5'-to-3' degradation of the DNA end is markedly reduced. The tetracycline-controlled depletion of Sth1, the ATPase of RSC, or deletion of RSC2 severely reduces chromatin remodeling and loading of Mre11 and Yku proteins at the DSB. Depletion of Sth1 also reduces phosphorylation of H2A, processing, and joining of DSBs. We propose that RSC-mediated chromatin remodeling at the DSB prepares chromatin to allow repair machinery to access the break and is vital for efficient DSB repair.
Figures
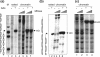
Similar articles
-
The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks.Mol Cell Biol. 2005 May;25(10):3934-44. doi: 10.1128/MCB.25.10.3934-3944.2005. Mol Cell Biol. 2005. PMID: 15870268 Free PMC article.
-
RSC functions as an early double-strand-break sensor in the cell's response to DNA damage.Curr Biol. 2007 Aug 21;17(16):1432-7. doi: 10.1016/j.cub.2007.07.035. Epub 2007 Aug 9. Curr Biol. 2007. PMID: 17689960 Free PMC article.
-
Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks.EMBO J. 2007 Sep 19;26(18):4113-25. doi: 10.1038/sj.emboj.7601835. Epub 2007 Aug 30. EMBO J. 2007. PMID: 17762868 Free PMC article.
-
Altering nucleosomes during DNA double-strand break repair in yeast.Trends Genet. 2006 Dec;22(12):671-7. doi: 10.1016/j.tig.2006.09.007. Epub 2006 Sep 25. Trends Genet. 2006. PMID: 16997415 Review.
-
The Chromatin Landscape around DNA Double-Strand Breaks in Yeast and Its Influence on DNA Repair Pathway Choice.Int J Mol Sci. 2023 Feb 7;24(4):3248. doi: 10.3390/ijms24043248. Int J Mol Sci. 2023. PMID: 36834658 Free PMC article. Review.
Cited by
-
Sequence and chromatin features guide DNA double-strand break resection initiation.Mol Cell. 2023 Apr 20;83(8):1237-1250.e15. doi: 10.1016/j.molcel.2023.02.010. Epub 2023 Mar 13. Mol Cell. 2023. PMID: 36917982 Free PMC article.
-
Features of cryptic promoters and their varied reliance on bromodomain-containing factors.PLoS One. 2010 Sep 23;5(9):e12927. doi: 10.1371/journal.pone.0012927. PLoS One. 2010. PMID: 20886085 Free PMC article.
-
Regulation of DNA Double Strand Breaks Processing: Focus on Barriers.Front Mol Biosci. 2019 Jul 16;6:55. doi: 10.3389/fmolb.2019.00055. eCollection 2019. Front Mol Biosci. 2019. PMID: 31380392 Free PMC article. Review.
-
Chromatin remodelling complex RSC promotes base excision repair in chromatin of Saccharomyces cerevisiae.DNA Repair (Amst). 2014 Apr;16:35-43. doi: 10.1016/j.dnarep.2014.01.002. Epub 2014 Feb 25. DNA Repair (Amst). 2014. PMID: 24674626 Free PMC article.
-
Histone H3 lysine 14 (H3K14) acetylation facilitates DNA repair in a positioned nucleosome by stabilizing the binding of the chromatin Remodeler RSC (Remodels Structure of Chromatin).J Biol Chem. 2014 Mar 21;289(12):8353-63. doi: 10.1074/jbc.M113.540732. Epub 2014 Feb 10. J Biol Chem. 2014. PMID: 24515106 Free PMC article.
References
-
- Ai, X., and M. R. Parthun. 2004. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14:195-205. - PubMed
-
- Bassing, C. H., K. F. Chua, J. Sekiguchi, H. Suh, S. R. Whitlow, J. C. Fleming, B. C. Monroe, D. N. Ciccone, C. Yan, K. Vlasakova, D. M. Livingston, D. O. Ferguson, R. Scully, and F. W. Alt. 2002. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA 99:8173-8178. - PMC - PubMed
-
- Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. - PubMed
-
- Cairns, B. R. 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15:185-190. - PubMed
-
- Celeste, A., S. Petersen, P. J. Romanienko, O. Fernandez-Capetillo, H. T. Chen, O. A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M. J. Difilippantonio, C. Redon, D. R. Pilch, A. Olaru, M. Eckhaus, R. D. Camerini-Otero, L. Tessarollo, F. Livak, K. Manova, W. M. Bonner, M. C. Nussenzweig, and A. Nussenzweig. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922-927. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
