Requirement of Nhp6 proteins for transcription of a subset of tRNA genes and heterochromatin barrier function in Saccharomyces cerevisiae
- PMID: 17178828
- PMCID: PMC1820459
- DOI: 10.1128/MCB.00773-06
Requirement of Nhp6 proteins for transcription of a subset of tRNA genes and heterochromatin barrier function in Saccharomyces cerevisiae
Abstract
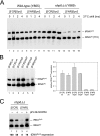
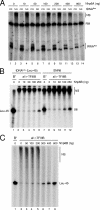
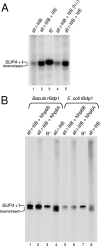
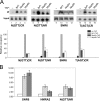
A key event in tRNA gene (tDNA) transcription by RNA polymerase (Pol) III is the TFIIIC-dependent assembly of TFIIIB upstream of the transcription start site. Different tDNA upstream sequences bind TFIIIB with different affinities, thereby modulating tDNA transcription. We found that in the absence of Nhp6 proteins, the influence of the 5'-flanking region on tRNA gene transcription is dramatically enhanced in Saccharomyces cerevisiae. Expression of a tDNA bearing a suboptimal TFIIIB binding site, but not of a tDNA preceded by a strong TFIIIB binding region, was strongly dependent on Nhp6 in vivo. Upstream sequence-dependent stimulation of tRNA gene transcription by Nhp6 could be reproduced in vitro, and Nhp6 proteins were found associated with tRNA genes in yeast cells. We also show that both transcription and silencing barrier activity of a tDNA(Thr) at the HMR locus are compromised in the absence of Nhp6. Our data suggest that Nhp6 proteins are important components of Pol III chromatin templates that contribute both to the robustness of tRNA gene expression and to positional effects of Pol III transcription complexes.
Figures


Similar articles
-
Nhp6 is a transcriptional initiation fidelity factor for RNA polymerase III transcription in vitro and in vivo.J Biol Chem. 2006 Mar 17;281(11):7445-51. doi: 10.1074/jbc.M512810200. Epub 2006 Jan 11. J Biol Chem. 2006. PMID: 16407207
-
TFIIIC-independent in vitro transcription of yeast tRNA genes.J Mol Biol. 2000 Jun 9;299(3):601-13. doi: 10.1006/jmbi.2000.3783. J Mol Biol. 2000. PMID: 10835271
-
Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae.Mol Cell. 2001 Feb;7(2):309-18. doi: 10.1016/s1097-2765(01)00179-4. Mol Cell. 2001. PMID: 11239460
-
Regulation of tRNA synthesis by the general transcription factors of RNA polymerase III - TFIIIB and TFIIIC, and by the MAF1 protein.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):320-329. doi: 10.1016/j.bbagrm.2018.01.011. Epub 2018 Feb 6. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29378333 Review.
-
Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human.Nucleic Acids Res. 2001 Jul 1;29(13):2675-90. doi: 10.1093/nar/29.13.2675. Nucleic Acids Res. 2001. PMID: 11433012 Free PMC article. Review.
Cited by
-
Regulation of pol III transcription by nutrient and stress signaling pathways.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):361-75. doi: 10.1016/j.bbagrm.2012.11.001. Epub 2012 Nov 16. Biochim Biophys Acta. 2013. PMID: 23165150 Free PMC article. Review.
-
Intergenic transcriptional interference is blocked by RNA polymerase III transcription factor TFIIIB in Saccharomyces cerevisiae.Genetics. 2014 Feb;196(2):427-38. doi: 10.1534/genetics.113.160093. Epub 2013 Dec 13. Genetics. 2014. PMID: 24336746 Free PMC article.
-
The life of U6 small nuclear RNA, from cradle to grave.RNA. 2018 Apr;24(4):437-460. doi: 10.1261/rna.065136.117. Epub 2018 Jan 24. RNA. 2018. PMID: 29367453 Free PMC article. Review.
-
An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1.PLoS One. 2007 Oct 31;2(10):e1099. doi: 10.1371/journal.pone.0001099. PLoS One. 2007. PMID: 17971862 Free PMC article.
-
The choreography of chromatin in RNA polymerase III regulation.Biochem Soc Trans. 2024 Jun 26;52(3):1173-1189. doi: 10.1042/BST20230770. Biochem Soc Trans. 2024. PMID: 38666598 Free PMC article. Review.
References
-
- Andrau, J. C., and M. Werner. 2001. B"-associated factor(s) involved in RNA polymerase III preinitiation complex formation and start-site selection. Eur. J. Biochem. 268:5167-5175. - PubMed
-
- Bianchi, M. E., and A. Agresti. 2005. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 15:496-506. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
