Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks
- PMID: 17173371
- PMCID: PMC2542901
- DOI: 10.1021/tx060164e
Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks
Abstract
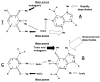
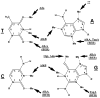
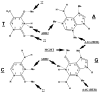
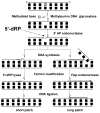
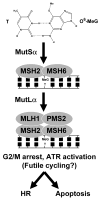
The chemical methylating agents methylmethane sulfonate (MMS) and N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) have been used for decades as classical DNA damaging agents. These agents have been utilized to uncover and explore pathways of DNA repair, DNA damage response, and mutagenesis. MMS and MNNG modify DNA by adding methyl groups to a number of nucleophilic sites on the DNA bases, although MNNG produces a greater percentage of O-methyl adducts. There has been substantial progress elucidating direct reversal proteins that remove methyl groups and base excision repair (BER), which removes and replaces methylated bases. Direct reversal proteins and BER, thus, counteract the toxic, mutagenic, and clastogenic effects of methylating agents. Despite recent progress, the complexity of DNA damage responses to methylating agents is still being discovered. In particular, there is growing understanding of pathways such as homologous recombination, lesion bypass, and mismatch repair that react when the response of direct reversal proteins and BER is insufficient. Furthermore, the importance of proper balance within the steps in BER has been uncovered with the knowledge that DNA structural intermediates during BER are deleterious. A number of issues complicate the elucidation of the downstream responses when direct reversal is insufficient or BER is imbalanced. These include inter-species differences, cell-type-specific differences within mammals and between cancer cell lines, and the type of methyl damage or BER intermediate encountered. MMS also carries a misleading reputation of being a radiomimetic, that is, capable of directly producing strand breaks. This review focuses on the DNA methyl damage caused by MMS and MNNG for each site of potential methylation to summarize what is known about the repair of such damage and the downstream responses and consequences if the damage is not repaired.
Figures
Similar articles
-
Homologous recombination prevents methylation-induced toxicity in Escherichia coli.Nucleic Acids Res. 2006 May 2;34(8):2258-68. doi: 10.1093/nar/gkl222. Print 2006. Nucleic Acids Res. 2006. PMID: 16670432 Free PMC article.
-
Homologous recombination protects mammalian cells from replication-associated DNA double-strand breaks arising in response to methyl methanesulfonate.DNA Repair (Amst). 2010 Oct 5;9(10):1050-63. doi: 10.1016/j.dnarep.2010.07.005. Epub 2010 Aug 13. DNA Repair (Amst). 2010. PMID: 20708982
-
Potentiation of cytotoxicity by 3-aminobenzamide in DNA repair-deficient human tumor cell lines following exposure to methylating agents or anti-neoplastic drugs.Carcinogenesis. 1988 Apr;9(4):541-6. doi: 10.1093/carcin/9.4.541. Carcinogenesis. 1988. PMID: 3356063
-
Mechanisms and consequences of methylating agent-induced SCEs and chromosomal aberrations: a long road traveled and still a far way to go.Cytogenet Genome Res. 2004;104(1-4):77-86. doi: 10.1159/000077469. Cytogenet Genome Res. 2004. PMID: 15162018 Review.
-
Biological Basis for Threshold Responses to Methylating Agents.Chem Res Toxicol. 2020 Sep 21;33(9):2219-2224. doi: 10.1021/acs.chemrestox.0c00052. Epub 2020 May 27. Chem Res Toxicol. 2020. PMID: 32388971 Free PMC article. Review.
Cited by
-
Structural and Dynamical Properties of Nucleic Acid Hairpins Implicated in Trinucleotide Repeat Expansion Diseases.Biomolecules. 2024 Oct 10;14(10):1278. doi: 10.3390/biom14101278. Biomolecules. 2024. PMID: 39456210 Free PMC article. Review.
-
ALKBH1 is dispensable for abasic site cleavage during base excision repair and class switch recombination.PLoS One. 2013 Jun 25;8(6):e67403. doi: 10.1371/journal.pone.0067403. Print 2013. PLoS One. 2013. PMID: 23825659 Free PMC article.
-
Hoogsteen base pairs increase the susceptibility of double-stranded DNA to cytotoxic damage.J Biol Chem. 2020 Nov 20;295(47):15933-15947. doi: 10.1074/jbc.RA120.014530. Epub 2020 Sep 10. J Biol Chem. 2020. PMID: 32913127 Free PMC article.
-
DNA Damage Response and DNA Repair in Skeletal Myocytes From a Mouse Model of Spinal Muscular Atrophy.J Neuropathol Exp Neurol. 2016 Sep;75(9):889-902. doi: 10.1093/jnen/nlw064. Epub 2016 Jul 24. J Neuropathol Exp Neurol. 2016. PMID: 27452406 Free PMC article.
-
Against the Resilience of High-Grade Gliomas: The Immunotherapeutic Approach (Part I).Brain Sci. 2021 Mar 18;11(3):386. doi: 10.3390/brainsci11030386. Brain Sci. 2021. PMID: 33803885 Free PMC article. Review.
References
-
- Pullman A, Pullman B. Molecular electrostatic potential of the nucleic acids. Q Rev Biophys. 1981;14:289–380. - PubMed
-
- Galtress CL, Morrow PR, Nag S, Smalley TL, Tschantz MF, Vaughn JS, Wichems DN, Ziglar SK, Fishbein JC. Mechanism for the Solvolytic Decomposition of the Carcinogen N-Methyl-N′-Nitro-N-Nitrosoguanidine in Aqueous-Solutions. J Am Chem Soc. 1992;114:1406–1411.
-
- Loechler EL. A violation of the Swain-Scott principle, and not SN1 versus SN2 reaction mechanisms, explains why carcinogenic alkylating agents can form different proportions of adducts at oxygen versus nitrogen in DNA. Chem Res Toxicol. 1994;7:277–280. - PubMed
-
- Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat Rev. 1997;23:35–61. - PubMed
-
- Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

