Proteasome substrate degradation requires association plus extended peptide
- PMID: 17170706
- PMCID: PMC1782366
- DOI: 10.1038/sj.emboj.7601476
Proteasome substrate degradation requires association plus extended peptide
Abstract
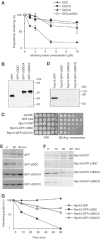
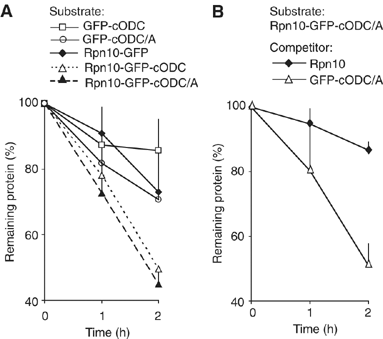
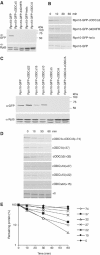
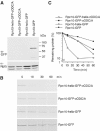
To determine the minimum requirements for substrate recognition and processing by proteasomes, the functional elements of a ubiquitin-independent degradation tag were dissected. The 37-residue C-terminus of ornithine decarboxylase (cODC) is a native degron, which also functions when appended to diverse proteins. Mutating the cysteine 441 residue within cODC impaired its proteasome association in the context of ornithine decarboxylase and prevented the turnover of GFP-cODC in yeast cells. Degradation of GFP-cODC with C441 mutations was restored by providing an alternate proteasome association element via fusion to the Rpn10 proteasome subunit. However, Rpn10-GFP was stable, unless extended by cODC or other peptides of similar size. In vitro reconstitution experiments confirmed the requirement for both proteasome tethering and a loosely structured region. Therefore, cODC and degradation tags in general must serve two functions: proteasome association and a site, consisting of an extended peptide region, used for initiating insertion into the protease.
Figures

Similar articles
-
Structural elements of the ubiquitin-independent proteasome degron of ornithine decarboxylase.Biochem J. 2008 Mar 1;410(2):401-7. doi: 10.1042/BJ20071239. Biochem J. 2008. PMID: 17979831
-
Proteasomes begin ornithine decarboxylase digestion at the C terminus.J Biol Chem. 2004 May 14;279(20):20959-65. doi: 10.1074/jbc.M314043200. Epub 2004 Mar 11. J Biol Chem. 2004. PMID: 15016805
-
Recognition of Poly-Ubiquitins by the Proteasome through Protein Refolding Guided by Electrostatic and Hydrophobic Interactions.J Phys Chem B. 2016 Aug 25;120(33):8137-46. doi: 10.1021/acs.jpcb.6b01327. Epub 2016 Apr 6. J Phys Chem B. 2016. PMID: 27012670 Free PMC article.
-
Targeting proteins for degradation.Nat Chem Biol. 2009 Nov;5(11):815-22. doi: 10.1038/nchembio.250. Nat Chem Biol. 2009. PMID: 19841631 Free PMC article. Review.
-
Degradation of ornithine decarboxylase by the 26S proteasome.Biochem Biophys Res Commun. 2000 Jan 7;267(1):1-6. doi: 10.1006/bbrc.1999.1706. Biochem Biophys Res Commun. 2000. PMID: 10623564 Review.
Cited by
-
Ubiquitin-independent proteasomal degradation.Biochim Biophys Acta. 2014 Jan;1843(1):216-21. doi: 10.1016/j.bbamcr.2013.05.008. Epub 2013 May 14. Biochim Biophys Acta. 2014. PMID: 23684952 Free PMC article. Review.
-
Targeted protein depletion in Saccharomyces cerevisiae by activation of a bidirectional degron.BMC Syst Biol. 2010 Dec 29;4:176. doi: 10.1186/1752-0509-4-176. BMC Syst Biol. 2010. PMID: 21190544 Free PMC article.
-
Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs).Chem Rev. 2014 Jul 9;114(13):6661-714. doi: 10.1021/cr400695p. Epub 2014 Jun 5. Chem Rev. 2014. PMID: 24901537 Free PMC article. Review. No abstract available.
-
Functional asymmetries of proteasome translocase pore.J Biol Chem. 2012 May 25;287(22):18535-43. doi: 10.1074/jbc.M112.357327. Epub 2012 Apr 5. J Biol Chem. 2012. PMID: 22493437 Free PMC article.
-
Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs.J Biol Chem. 2013 Mar 15;288(11):7781-7790. doi: 10.1074/jbc.M112.441907. Epub 2013 Jan 22. J Biol Chem. 2013. PMID: 23341450 Free PMC article.
References
-
- Carrion-Vazquez M, Oberhauser AF, Fisher TE, Marszalek PE, Li H, Fernandez JM (2000) Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog Biophys Mol Biol 74: 63–91 - PubMed
-
- Chen H, MacDonald A, Coffino P (2002) Structural elements of antizymes 1 and 2 required for proteasomal degradation of ornithine decarboxylase. J Biol Chem 277: 45957–45961 - PubMed
-
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATà2 repressor. Cell 74: 357–369 - PubMed
-
- Ghoda L, van Daalen Wetters T, Macrae M, Ascherman D, Coffino P (1989) Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science 243: 1493–1495 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

