The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis
- PMID: 17159899
- PMCID: PMC1782375
- DOI: 10.1038/sj.emboj.7601478
The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis
Abstract
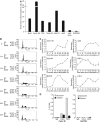
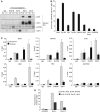
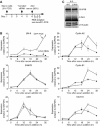
Regulated gene expression is critical for the proper timing of cell cycle transitions. Here we report that human LIN-9 has an important function in transcriptional regulation of G2/M genes. Depletion of LIN-9 by RNAi in human fibroblasts strongly impairs proliferation and delays progression from G2 to M. We identify a cluster of G2/M genes as direct targets of LIN-9. Activation of these genes is linked to an association between LIN-9 and B-MYB. Chromatin immunoprecipitation assays revealed binding of both LIN-9 and B-MYB to the promoters of G2/M regulated genes. Depletion of B-MYB recapitulated the biological outcome of LIN-9 knockdown, including impaired proliferation and reduced expression of G2/M genes. These data suggest a critical role for human LIN-9, together with B-MYB, in the activation of genes that are essential for progression into mitosis.
Figures


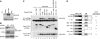
Similar articles
-
A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells.Oncogene. 2009 Apr 16;28(15):1737-47. doi: 10.1038/onc.2009.22. Epub 2009 Mar 2. Oncogene. 2009. PMID: 19252525
-
B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells.Cancer Res. 2009 May 1;69(9):4073-80. doi: 10.1158/0008-5472.CAN-08-4156. Epub 2009 Apr 21. Cancer Res. 2009. PMID: 19383908
-
Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters.Nature. 2000 Jul 6;406(6791):94-8. doi: 10.1038/35017589. Nature. 2000. PMID: 10894549
-
B-Myb protein in cellular proliferation, transcription control, and cancer: latest developments.J Cell Physiol. 1999 Jun;179(3):245-50. doi: 10.1002/(SICI)1097-4652(199906)179:3<245::AID-JCP1>3.0.CO;2-H. J Cell Physiol. 1999. PMID: 10228942 Review.
-
[Regulation of plant cell cycle by Myb-like transcription factors].Tanpakushitsu Kakusan Koso. 2002 Sep;47(12 Suppl):1633-8. Tanpakushitsu Kakusan Koso. 2002. PMID: 12357627 Review. Japanese. No abstract available.
Cited by
-
p53 can repress transcription of cell cycle genes through a p21(WAF1/CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements.Cell Cycle. 2012 Dec 15;11(24):4661-72. doi: 10.4161/cc.22917. Epub 2012 Nov 27. Cell Cycle. 2012. PMID: 23187802 Free PMC article.
-
p53 and cell cycle dependent transcription of kinesin family member 23 (KIF23) is controlled via a CHR promoter element bound by DREAM and MMB complexes.PLoS One. 2013 May 1;8(5):e63187. doi: 10.1371/journal.pone.0063187. Print 2013. PLoS One. 2013. PMID: 23650552 Free PMC article.
-
The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression.Genes Dev. 2012 Mar 1;26(5):474-89. doi: 10.1101/gad.181933.111. Genes Dev. 2012. PMID: 22391450 Free PMC article.
-
Expression of the cytokinesis regulator PRC1 results in p53-pathway activation in A549 cells but does not directly regulate gene expression in the nucleus.Cell Cycle. 2023 Feb;22(4):419-432. doi: 10.1080/15384101.2022.2122258. Epub 2022 Sep 22. Cell Cycle. 2023. PMID: 36135961 Free PMC article.
-
Upregulation of the tumor suppressor gene LIN9 enhances tumorigenesis and predicts poor prognosis of lung adenocarcinoma.Heliyon. 2024 Jul 24;10(15):e35012. doi: 10.1016/j.heliyon.2024.e35012. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39157309 Free PMC article.
References
-
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833–837 - PubMed
-
- Begg AC, McNally NJ, Shrieve DC, Karcher H (1985) A method to measure the duration of DNA synthesis and the potential doubling time from a single sample. Cytometry 6: 620–626 - PubMed
-
- Carvalho A, Carmena M, Sambade C, Earnshaw WC, Wheatley SP (2003) Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J Cell Sci 116: 2987–2998 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

