Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense
- PMID: 17158744
- PMCID: PMC1686603
- DOI: 10.1101/gad.1495506
Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense
Abstract
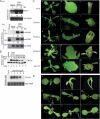
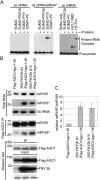
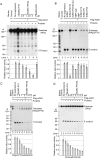
RNA silencing refers to small regulatory RNA-mediated processes that repress endogenous gene expression and defend hosts from offending viruses. As an anti-host defense mechanism, viruses encode suppressors that can block RNA silencing pathways. Cucumber mosaic virus (CMV)-encoded 2b protein was among the first suppressors identified that could inhibit post-transcriptional gene silencing (PTGS), but with little or no effect on miRNA functions. The mechanisms underlying 2b suppression of RNA silencing are unknown. Here, we demonstrate that the CMV 2b protein also interferes with miRNA pathways, eliciting developmental anomalies partially phenocopying ago1 mutant alleles. In contrast to most characterized suppressors, 2b directly interacts with Argonaute1 (AGO1) in vitro and in vivo, and this interaction occurs primarily on one surface of the PAZ-containing module and part of the PIWI-box of AGO1. Consistent with this interaction, 2b specifically inhibits AGO1 cleavage activity in RISC reconstitution assays. In addition, AGO1 recruits virus-derived small interfering RNAs (siRNAs) in vivo, suggesting that AGO1 is a major factor in defense against CMV infection. We conclude that 2b blocks AGO1 cleavage activity to inhibit miRNA pathways, attenuate RNA silencing, and counter host defense. These findings provide insight on the molecular arms race between host antiviral RNA silencing and virus counterdefense.
Figures




Similar articles
-
Strain-specific differences in the interactions of the cucumber mosaic virus 2b protein with the viral 1a and host Argonaute 1 proteins.J Virol. 2024 Sep 17;98(9):e0099324. doi: 10.1128/jvi.00993-24. Epub 2024 Aug 20. J Virol. 2024. PMID: 39162432
-
Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities.Plant J. 2012 Jan;69(1):104-15. doi: 10.1111/j.1365-313X.2011.04774.x. Epub 2011 Nov 8. Plant J. 2012. PMID: 21880078
-
The cucumber mosaic virus 1a protein regulates interactions between the 2b protein and ARGONAUTE 1 while maintaining the silencing suppressor activity of the 2b protein.PLoS Pathog. 2020 Dec 3;16(12):e1009125. doi: 10.1371/journal.ppat.1009125. eCollection 2020 Dec. PLoS Pathog. 2020. PMID: 33270799 Free PMC article.
-
[The battle of Silence : action and inhibition of RNA silencing during plant/virus interactions].Med Sci (Paris). 2009 May;25(5):505-11. doi: 10.1051/medsci/2009255505. Med Sci (Paris). 2009. PMID: 19480832 Review. French.
-
Structural insights into the arms race between host and virus along RNA silencing pathways in Arabidopsis thaliana.Biol Rev Camb Philos Soc. 2014 May;89(2):337-55. doi: 10.1111/brv.12057. Epub 2013 Sep 4. Biol Rev Camb Philos Soc. 2014. PMID: 24034134 Review.
Cited by
-
Antiviral roles of plant ARGONAUTES.Curr Opin Plant Biol. 2015 Oct;27:111-7. doi: 10.1016/j.pbi.2015.06.013. Epub 2015 Jul 17. Curr Opin Plant Biol. 2015. PMID: 26190744 Free PMC article. Review.
-
Predicted trans-acting siRNAs in the human brain.Int J Mol Sci. 2015 Feb 3;16(2):3377-90. doi: 10.3390/ijms16023377. Int J Mol Sci. 2015. PMID: 25654231 Free PMC article.
-
Regulation of microRNA biogenesis and turnover by animals and their viruses.Cell Mol Life Sci. 2013 Oct;70(19):3525-44. doi: 10.1007/s00018-012-1257-1. Epub 2013 Jan 26. Cell Mol Life Sci. 2013. PMID: 23354060 Free PMC article. Review.
-
The Functions of RNA-Dependent RNA Polymerases in Arabidopsis.Arabidopsis Book. 2011;9:e0146. doi: 10.1199/tab.0146. Epub 2011 Jul 31. Arabidopsis Book. 2011. PMID: 22303271 Free PMC article.
-
Contribution of small RNA pathway components in plant immunity.Mol Plant Microbe Interact. 2013 Jun;26(6):617-25. doi: 10.1094/MPMI-10-12-0255-IA. Mol Plant Microbe Interact. 2013. PMID: 23489060 Free PMC article. Review.
References
-
- Allen, E., Xie, Z.X., Gustafson, A., Carrington, J. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. - PubMed
-
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Baulcombe, D. RNA silencing in plants. Nature. 2004;431:356–363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
