Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development
- PMID: 17158604
- PMCID: PMC1785416
- DOI: 10.1105/tpc.106.047605
Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development
Abstract
mRNA turnover in eukaryotes involves the removal of m7GDP from the 5' end. This decapping reaction is mediated by a protein complex well characterized in yeast and human but not in plants. The function of the decapping complex in the development of multicellular organisms is also poorly understood. Here, we show that Arabidopsis thaliana DCP2 can generate from capped mRNAs, m7GDP, and 5'-phosphorylated mRNAs in vitro and that this decapping activity requires an active Nudix domain. DCP2 interacts in vitro and in vivo with DCP1 and VARICOSE (VCS), an Arabidopsis homolog of human Hedls/Ge-1. Moreover, the interacting proteins stimulate DCP2 activity, suggesting that the three proteins operate as a decapping complex. Consistent with their role in mRNA decay, DCP1, DCP2, and VCS colocalize in cytoplasmic foci, which are putative Arabidopsis processing bodies. Compared with the wild type, null mutants of DCP1, DCP2, and VCS accumulate capped mRNAs with a reduced degradation rate. These mutants also share a similar lethal phenotype at the seedling cotyledon stage, with disorganized veins, swollen root hairs, and altered epidermal cell morphology. We conclude that mRNA turnover mediated by the decapping complex is required for postembryonic development in Arabidopsis.
Figures

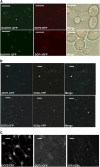

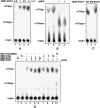



Similar articles
-
The role of decapping proteins in the miRNA accumulation in Arabidopsis thaliana.RNA Biol. 2012 May;9(5):644-52. doi: 10.4161/rna.19877. Epub 2012 May 1. RNA Biol. 2012. PMID: 22614834
-
New insights into the control of mRNA decapping.Trends Biochem Sci. 2006 May;31(5):241-3. doi: 10.1016/j.tibs.2006.03.001. Epub 2006 Mar 31. Trends Biochem Sci. 2006. PMID: 16580207 Review.
-
The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis.Plant J. 2012 Nov;72(3):368-77. doi: 10.1111/j.1365-313X.2012.05066.x. Epub 2012 Aug 30. Plant J. 2012. PMID: 22639932
-
Diffuse decapping enzyme DCP2 accumulates in DCP1 foci under heat stress in Arabidopsis thaliana.Plant Cell Physiol. 2015 Jan;56(1):107-15. doi: 10.1093/pcp/pcu151. Epub 2014 Oct 22. Plant Cell Physiol. 2015. PMID: 25339350
-
RNA decapping inside and outside of processing bodies.Curr Opin Cell Biol. 2005 Jun;17(3):326-31. doi: 10.1016/j.ceb.2005.04.002. Curr Opin Cell Biol. 2005. PMID: 15901504 Review.
Cited by
-
A Forward Genetic Approach to Mapping a P-Element Second Site Mutation Identifies DCP2 as a Novel Tumor Suppressor in Drosophila melanogaster.G3 (Bethesda). 2020 Aug 5;10(8):2601-2618. doi: 10.1534/g3.120.401501. G3 (Bethesda). 2020. PMID: 32591349 Free PMC article.
-
Exploring Breakthroughs in Three Traits Belonging to Seed Life.Plants (Basel). 2022 Feb 11;11(4):490. doi: 10.3390/plants11040490. Plants (Basel). 2022. PMID: 35214823 Free PMC article. Review.
-
INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis.Plant Cell. 2007 Jun;19(6):1885-97. doi: 10.1105/tpc.106.045666. Epub 2007 Jun 29. Plant Cell. 2007. PMID: 17601829 Free PMC article.
-
The IκB kinase complex is a regulator of mRNA stability.EMBO J. 2018 Dec 14;37(24):e98658. doi: 10.15252/embj.201798658. Epub 2018 Nov 22. EMBO J. 2018. PMID: 30467221 Free PMC article.
-
PpNUDX8, a Peach NUDIX Hydrolase, Plays a Negative Regulator in Response to Drought Stress.Front Plant Sci. 2022 Feb 16;12:831883. doi: 10.3389/fpls.2021.831883. eCollection 2021. Front Plant Sci. 2022. PMID: 35251068 Free PMC article.
References
-
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., and Parker, R. (1996). An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382 642–646. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

