Ensembl 2007
- PMID: 17148474
- PMCID: PMC1761443
- DOI: 10.1093/nar/gkl996
Ensembl 2007
Abstract
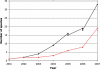
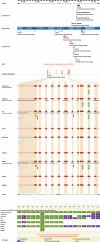
The Ensembl (http://www.ensembl.org/) project provides a comprehensive and integrated source of annotation of chordate genome sequences. Over the past year the number of genomes available from Ensembl has increased from 15 to 33, with the addition of sites for the mammalian genomes of elephant, rabbit, armadillo, tenrec, platypus, pig, cat, bush baby, common shrew, microbat and european hedgehog; the fish genomes of stickleback and medaka and the second example of the genomes of the sea squirt (Ciona savignyi) and the mosquito (Aedes aegypti). Some of the major features added during the year include the first complete gene sets for genomes with low-sequence coverage, the introduction of new strain variation data and the introduction of new orthology/paralog annotations based on gene trees.
Figures



Similar articles
-
Ensembl 2006.Nucleic Acids Res. 2006 Jan 1;34(Database issue):D556-61. doi: 10.1093/nar/gkj133. Nucleic Acids Res. 2006. PMID: 16381931 Free PMC article.
-
Ensembl 2005.Nucleic Acids Res. 2005 Jan 1;33(Database issue):D447-53. doi: 10.1093/nar/gki138. Nucleic Acids Res. 2005. PMID: 15608235 Free PMC article.
-
Ensembl 2009.Nucleic Acids Res. 2009 Jan;37(Database issue):D690-7. doi: 10.1093/nar/gkn828. Epub 2008 Nov 25. Nucleic Acids Res. 2009. PMID: 19033362 Free PMC article.
-
Ensembl 2008.Nucleic Acids Res. 2008 Jan;36(Database issue):D707-14. doi: 10.1093/nar/gkm988. Epub 2007 Nov 13. Nucleic Acids Res. 2008. PMID: 18000006 Free PMC article.
-
An overview of Ensembl.Genome Res. 2004 May;14(5):925-8. doi: 10.1101/gr.1860604. Epub 2004 Apr 12. Genome Res. 2004. PMID: 15078858 Free PMC article. Review.
Cited by
-
CREDO: a structural interactomics database for drug discovery.Database (Oxford). 2013 Jul 18;2013:bat049. doi: 10.1093/database/bat049. Print 2013. Database (Oxford). 2013. PMID: 23868908 Free PMC article.
-
Nucleosomes are well positioned in exons and carry characteristic histone modifications.Genome Res. 2009 Oct;19(10):1732-41. doi: 10.1101/gr.092353.109. Epub 2009 Aug 17. Genome Res. 2009. PMID: 19687145 Free PMC article.
-
Computational analysis of candidate disease genes and variants for salt-sensitive hypertension in indigenous Southern Africans.PLoS One. 2010 Sep 27;5(9):e12989. doi: 10.1371/journal.pone.0012989. PLoS One. 2010. PMID: 20886000 Free PMC article.
-
WormBase 2007.Nucleic Acids Res. 2008 Jan;36(Database issue):D612-7. doi: 10.1093/nar/gkm975. Epub 2007 Nov 8. Nucleic Acids Res. 2008. PMID: 17991679 Free PMC article.
-
Alternative splicing and protein structure evolution.Nucleic Acids Res. 2008 Feb;36(2):550-8. doi: 10.1093/nar/gkm1054. Epub 2007 Nov 30. Nucleic Acids Res. 2008. PMID: 18055499 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- BBS/B/13462/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/13446/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E010768/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/13470/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/13438/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

