Stochastic processes are key determinants of short-term evolution in influenza a virus
- PMID: 17140286
- PMCID: PMC1665651
- DOI: 10.1371/journal.ppat.0020125
Stochastic processes are key determinants of short-term evolution in influenza a virus
Erratum in
- PLoS Pathog. 2006 Dec;2(12):e138. Ghedi, Elodie [corrected to Ghedin, Elodie]
Abstract
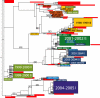
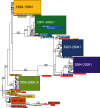
Understanding the evolutionary dynamics of influenza A virus is central to its surveillance and control. While immune-driven antigenic drift is a key determinant of viral evolution across epidemic seasons, the evolutionary processes shaping influenza virus diversity within seasons are less clear. Here we show with a phylogenetic analysis of 413 complete genomes of human H3N2 influenza A viruses collected between 1997 and 2005 from New York State, United States, that genetic diversity is both abundant and largely generated through the seasonal importation of multiple divergent clades of the same subtype. These clades cocirculated within New York State, allowing frequent reassortment and generating genome-wide diversity. However, relatively low levels of positive selection and genetic diversity were observed at amino acid sites considered important in antigenic drift. These results indicate that adaptive evolution occurs only sporadically in influenza A virus; rather, the stochastic processes of viral migration and clade reassortment play a vital role in shaping short-term evolutionary dynamics. Thus, predicting future patterns of influenza virus evolution for vaccine strain selection is inherently complex and requires intensive surveillance, whole-genome sequencing, and phenotypic analysis.
Conflict of interest statement
Figures
Similar articles
-
Molecular and antigenic evolution of human influenza A/H3N2 viruses in Quebec, Canada, 2009-2011.J Clin Virol. 2012 Jan;53(1):88-92. doi: 10.1016/j.jcv.2011.09.016. Epub 2011 Oct 28. J Clin Virol. 2012. PMID: 22036039
-
Molecular epidemiology and evolution of A(H1N1)pdm09 and H3N2 virus during winter 2012-2013 in Beijing, China.Infect Genet Evol. 2014 Aug;26:228-40. doi: 10.1016/j.meegid.2014.05.034. Epub 2014 Jun 7. Infect Genet Evol. 2014. PMID: 24911284
-
Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses.PLoS Biol. 2005 Sep;3(9):e300. doi: 10.1371/journal.pbio.0030300. Epub 2005 Jul 26. PLoS Biol. 2005. PMID: 16026181 Free PMC article.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
-
[Molecular characterization of human influenza viruses--a look back on the last 10 years].Berl Munch Tierarztl Wochenschr. 2006 Mar-Apr;119(3-4):167-78. Berl Munch Tierarztl Wochenschr. 2006. PMID: 16573207 Review. German.
Cited by
-
Long-term evolution of human seasonal influenza virus A(H3N2) is associated with an increase in polymerase complex activity.Virus Evol. 2024 May 4;10(1):veae030. doi: 10.1093/ve/veae030. eCollection 2024. Virus Evol. 2024. PMID: 38808037 Free PMC article.
-
Epidemiological and Virological Characterization of Influenza B Virus Infections.PLoS One. 2016 Aug 17;11(8):e0161195. doi: 10.1371/journal.pone.0161195. eCollection 2016. PLoS One. 2016. PMID: 27533045 Free PMC article.
-
Computational Characterization of Transient Strain-Transcending Immunity against Influenza A.PLoS One. 2015 May 1;10(5):e0125047. doi: 10.1371/journal.pone.0125047. eCollection 2015. PLoS One. 2015. PMID: 25933195 Free PMC article.
-
Contrasting the epidemiological and evolutionary dynamics of influenza spatial transmission.Philos Trans R Soc Lond B Biol Sci. 2013 Feb 4;368(1614):20120199. doi: 10.1098/rstb.2012.0199. Print 2013 Mar 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23382422 Free PMC article. Review.
-
Global migration dynamics underlie evolution and persistence of human influenza A (H3N2).PLoS Pathog. 2010 May 27;6(5):e1000918. doi: 10.1371/journal.ppat.1000918. PLoS Pathog. 2010. PMID: 20523898 Free PMC article.
References
-
- Bush RM, Fitch WM, Bender CA, Cox NJ. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol Biol Evol. 1999;16:1457–1465. - PubMed
-
- Bush RM Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science. 1999;286:1921–1925. - PubMed
-
- Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

