Atg27 is required for autophagy-dependent cycling of Atg9
- PMID: 17135291
- PMCID: PMC1783788
- DOI: 10.1091/mbc.e06-07-0612
Atg27 is required for autophagy-dependent cycling of Atg9
Abstract
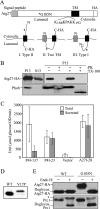
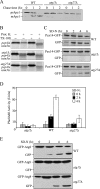
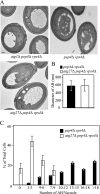
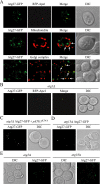
Autophagy is a catabolic pathway for the degradation of cytosolic proteins or organelles and is conserved among all eukaryotic cells. The hallmark of autophagy is the formation of double-membrane cytosolic vesicles, termed autophagosomes, which sequester cytoplasm; however, the mechanism of vesicle formation and the membrane source remain unclear. In the yeast Saccharomyces cerevisiae, selective autophagy mediates the delivery of specific cargos to the vacuole, the analog of the mammalian lysosome. The transmembrane protein Atg9 cycles between the mitochondria and the pre-autophagosomal structure, which is the site of autophagosome biogenesis. Atg9 is thought to mediate the delivery of membrane to the forming autophagosome. Here, we characterize a second transmembrane protein Atg27 that is required for specific autophagy in yeast. Atg27 is required for Atg9 cycling and shuttles between the pre-autophagosomal structure, mitochondria, and the Golgi complex. These data support a hypothesis that multiple membrane sources supply the lipids needed for autophagosome formation.
Figures
Similar articles
-
Atg27 is a second transmembrane cycling protein.Autophagy. 2007 May-Jun;3(3):254-6. doi: 10.4161/auto.3823. Epub 2007 May 11. Autophagy. 2007. PMID: 17297289
-
Atg27 tyrosine sorting motif is important for its trafficking and Atg9 localization.Traffic. 2015 Apr;16(4):365-78. doi: 10.1111/tra.12253. Epub 2015 Feb 20. Traffic. 2015. PMID: 25557545 Free PMC article.
-
Atg9 vesicles are an important membrane source during early steps of autophagosome formation.J Cell Biol. 2012 Jul 23;198(2):219-33. doi: 10.1083/jcb.201202061. J Cell Biol. 2012. PMID: 22826123 Free PMC article.
-
Atg9 trafficking in the yeast Saccharomyces cerevisiae.Autophagy. 2007 Mar-Apr;3(2):145-8. doi: 10.4161/auto.3608. Epub 2007 Mar 21. Autophagy. 2007. PMID: 17204846 Review.
-
Mechanisms of autophagosome biogenesis.Curr Biol. 2012 Jan 10;22(1):R29-34. doi: 10.1016/j.cub.2011.11.034. Curr Biol. 2012. PMID: 22240478 Review.
Cited by
-
The mechanism and physiological function of macroautophagy.J Innate Immun. 2013;5(5):427-33. doi: 10.1159/000351979. Epub 2013 Jun 11. J Innate Immun. 2013. PMID: 23774579 Free PMC article. Review.
-
ER-phagy: mechanisms, regulation, and diseases connected to the lysosomal clearance of the endoplasmic reticulum.Physiol Rev. 2022 Jul 1;102(3):1393-1448. doi: 10.1152/physrev.00038.2021. Epub 2022 Feb 21. Physiol Rev. 2022. PMID: 35188422 Free PMC article. Review.
-
Conserved and Diversified Mechanism of Autophagy between Plants and Animals upon Various Stresses.Antioxidants (Basel). 2021 Oct 29;10(11):1736. doi: 10.3390/antiox10111736. Antioxidants (Basel). 2021. PMID: 34829607 Free PMC article. Review.
-
The machinery of macroautophagy.Cell Res. 2014 Jan;24(1):24-41. doi: 10.1038/cr.2013.168. Epub 2013 Dec 24. Cell Res. 2014. PMID: 24366339 Free PMC article. Review.
-
Regulation of autophagy: modulation of the size and number of autophagosomes.FEBS Lett. 2014 Aug 1;588(15):2457-63. doi: 10.1016/j.febslet.2014.06.015. Epub 2014 Jun 10. FEBS Lett. 2014. PMID: 24928445 Free PMC article. Review.
References
-
- Campbell T. N., Choy F. Y. Expression of two green fluorescent protein variants in citrate-buffered media in Pichia pastoris. Anal. Biochem. 2002;311:193–195. - PubMed
-
- Dunn W. A., Jr, Cregg J. M., Kiel J.A.K.W., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Peoxphagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. - PubMed
-
- Gerhardt B., Kordas T. J., Thompson C. M., Patel P., Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J. Biol. Chem. 1998;273:15818–15829. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

