Vaccinia virus N1L protein resembles a B cell lymphoma-2 (Bcl-2) family protein
- PMID: 17123957
- PMCID: PMC2222835
- DOI: 10.1110/ps.062454707
Vaccinia virus N1L protein resembles a B cell lymphoma-2 (Bcl-2) family protein
Abstract
Poxviruses encode immuno-modulatory proteins capable of subverting host defenses. The poxvirus vaccinia expresses a small 14-kDa protein, N1L, that is critical for virulence. We report the crystal structure of N1L, which reveals an unexpected but striking resemblance to host apoptotic regulators of the B cell lymphoma-2 (Bcl-2) family. Although N1L lacks detectable Bcl-2 homology (BH) motifs at the sequence level, we show that N1L binds with high affinity to the BH3 peptides of pro-apoptotic Bcl-2 family proteins in vitro, consistent with a role for N1L in modulating host antiviral defenses.
Figures
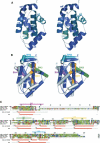
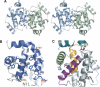
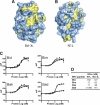
Comment in
-
The age of reverse biochemistry.Protein Sci. 2007 Jan;16(1):2-3. doi: 10.1110/ps.062615307. Protein Sci. 2007. PMID: 17192585 Free PMC article. No abstract available.
Similar articles
-
Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands.Cell Death Differ. 2008 Oct;15(10):1564-71. doi: 10.1038/cdd.2008.83. Epub 2008 Jun 13. Cell Death Differ. 2008. PMID: 18551131
-
Vaccinia virus protein A49 is an unexpected member of the B-cell Lymphoma (Bcl)-2 protein family.J Biol Chem. 2015 Mar 6;290(10):5991-6002. doi: 10.1074/jbc.M114.624650. Epub 2015 Jan 20. J Biol Chem. 2015. PMID: 25605733 Free PMC article.
-
Functional and structural studies of the vaccinia virus virulence factor N1 reveal a Bcl-2-like anti-apoptotic protein.J Gen Virol. 2007 Jun;88(Pt 6):1656-1666. doi: 10.1099/vir.0.82772-0. J Gen Virol. 2007. PMID: 17485524 Free PMC article.
-
Structural biology of the Bcl-2 family of proteins.Biochim Biophys Acta. 2004 Mar 1;1644(2-3):83-94. doi: 10.1016/j.bbamcr.2003.08.012. Biochim Biophys Acta. 2004. PMID: 14996493 Review.
-
Poxvirus antagonism of innate immunity by Bcl-2 fold proteins.J Struct Biol. 2013 Jan;181(1):1-10. doi: 10.1016/j.jsb.2012.10.015. Epub 2012 Nov 5. J Struct Biol. 2013. PMID: 23138003 Review.
Cited by
-
Caspase-dependent inhibition of mousepox replication by gzmB.PLoS One. 2009 Oct 19;4(10):e7512. doi: 10.1371/journal.pone.0007512. PLoS One. 2009. PMID: 19838298 Free PMC article.
-
The Bcl-2 Family: Ancient Origins, Conserved Structures, and Divergent Mechanisms.Biomolecules. 2020 Jan 12;10(1):128. doi: 10.3390/biom10010128. Biomolecules. 2020. PMID: 31940915 Free PMC article. Review.
-
Robust intrapulmonary CD8 T cell responses and protection with an attenuated N1L deleted vaccinia virus.PLoS One. 2008 Oct 2;3(10):e3323. doi: 10.1371/journal.pone.0003323. PLoS One. 2008. PMID: 18830408 Free PMC article.
-
How vaccinia virus has evolved to subvert the host immune response.J Struct Biol. 2011 Aug;175(2):127-34. doi: 10.1016/j.jsb.2011.03.010. Epub 2011 Mar 17. J Struct Biol. 2011. PMID: 21419849 Free PMC article. Review.
-
Deletion of the vaccinia virus N2L gene encoding an inhibitor of IRF3 improves the immunogenicity of modified vaccinia virus Ankara expressing HIV-1 antigens.J Virol. 2014 Mar;88(6):3392-410. doi: 10.1128/JVI.02723-13. Epub 2014 Jan 3. J Virol. 2014. PMID: 24390336 Free PMC article.
References
-
- Abrahams, J.P. and Leslie, A.G. 1996. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52: 30–42. - PubMed
-
- Antoine, G., Scheiflinger, F., Dorner, F., and Falkner, F.G. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: Comparison with other orthopoxviruses. Virology 244: 365–396. - PubMed
-
- Bartlett, N., Symons, J.A., Tscharke, D.C., and Smith, G.L. 2002. The vaccinia virus N1L protein is an intracellular homodimer that promotes virulence. J. Gen. Virol. 83: 1965–1976. - PubMed
-
- Bowie, A.G., Zhan, J., and Marshall, W.L. 2004. Viral appropriation of apoptotic and NF-κB signaling pathways. J. Cell. Biochem. 91: 1099–1108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

