Operon conservation and the evolution of trans-splicing in the phylum Nematoda
- PMID: 17121468
- PMCID: PMC1657053
- DOI: 10.1371/journal.pgen.0020198
Operon conservation and the evolution of trans-splicing in the phylum Nematoda
Abstract
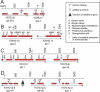
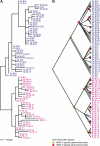
The nematode Caenorhabditis elegans is unique among model animals in that many of its genes are cotranscribed as polycistronic pre-mRNAs from operons. The mechanism by which these operonic transcripts are resolved into mature mRNAs includes trans-splicing to a family of SL2-like spliced leader exons. SL2-like spliced leaders are distinct from SL1, the major spliced leader in C. elegans and other nematode species. We surveyed five additional nematode species, representing three of the five major clades of the phylum Nematoda, for the presence of operons and the use of trans-spliced leaders in resolution of polycistronic pre-mRNAs. Conserved operons were found in Pristionchus pacificus, Nippostrongylus brasiliensis, Strongyloides ratti, Brugia malayi, and Ascaris suum. In nematodes closely related to the rhabditine C. elegans, a related family of SL2-like spliced leaders is used for operonic transcript resolution. However, in the tylenchine S. ratti operonic transcripts are resolved using a family of spliced leaders related to SL1. Non-operonic genes in S. ratti may also receive these SL1 variants. In the spirurine nematodes B. malayi and A. suum operonic transcripts are resolved using SL1. Mapping these phenotypes onto the robust molecular phylogeny for the Nematoda suggests that operons evolved before SL2-like spliced leaders, which are an evolutionary invention of the rhabditine lineage.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

Similar articles
-
Resolution of polycistronic RNA by SL2 trans-splicing is a widely conserved nematode trait.RNA. 2020 Dec;26(12):1891-1904. doi: 10.1261/rna.076414.120. Epub 2020 Sep 4. RNA. 2020. PMID: 32887788 Free PMC article.
-
SL2-like spliced leader RNAs in the basal nematode Prionchulus punctatus: New insight into the evolution of nematode SL2 RNAs.RNA. 2010 Aug;16(8):1500-7. doi: 10.1261/rna.2155010. Epub 2010 Jun 21. RNA. 2010. PMID: 20566669 Free PMC article.
-
Operon structure and trans-splicing in the nematode Pristionchus pacificus.Mol Biol Evol. 2003 Dec;20(12):2097-103. doi: 10.1093/molbev/msg225. Epub 2003 Aug 29. Mol Biol Evol. 2003. PMID: 12949121
-
The evolution of spliced leader trans-splicing in nematodes.Biochem Soc Trans. 2010 Aug;38(4):1125-30. doi: 10.1042/BST0381125. Biochem Soc Trans. 2010. PMID: 20659016 Review.
-
Trans-splicing and operons in C. elegans.WormBook. 2012 Nov 20:1-11. doi: 10.1895/wormbook.1.5.2. WormBook. 2012. PMID: 23175478 Free PMC article. Review.
Cited by
-
A novel family of C. elegans snRNPs contains proteins associated with trans-splicing.RNA. 2007 Apr;13(4):511-20. doi: 10.1261/rna.426707. Epub 2007 Feb 5. RNA. 2007. PMID: 17283210 Free PMC article.
-
Expression profiling and cross-species RNA interference (RNAi) of desiccation-induced transcripts in the anhydrobiotic nematode Aphelenchus avenae.BMC Mol Biol. 2010 Jan 19;11:6. doi: 10.1186/1471-2199-11-6. BMC Mol Biol. 2010. PMID: 20085654 Free PMC article.
-
Genome-wide analysis of trans-splicing in the nematode Pristionchus pacificus unravels conserved gene functions for germline and dauer development in divergent operons.RNA. 2014 Sep;20(9):1386-97. doi: 10.1261/rna.041954.113. Epub 2014 Jul 11. RNA. 2014. PMID: 25015138 Free PMC article.
-
C. elegans sequences that control trans-splicing and operon pre-mRNA processing.RNA. 2007 Sep;13(9):1409-26. doi: 10.1261/rna.596707. Epub 2007 Jul 13. RNA. 2007. PMID: 17630324 Free PMC article.
-
Metazoan operons accelerate recovery from growth-arrested states.Cell. 2011 Jun 10;145(6):981-92. doi: 10.1016/j.cell.2011.05.013. Cell. 2011. PMID: 21663799 Free PMC article.
References
-
- Bonen L. Trans-splicing of mRNA in plants, animals and protists. FASEB J. 1993;7:40–46. - PubMed
-
- Landfear SM, Wirth DF. Structure of mRNA encoded by tubulin genes in Leishmania enriettii . Mol Biochem Parasitol. 1985;15:61–82. - PubMed
-
- Kroiher M, Reidling JC, Steele RE. A gene whose major transcript encodes only the substrate-binding domain of a protein-tyrosine kinase. Gene. 2000;241:317–324. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

