Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus
- PMID: 17114349
- PMCID: PMC1693932
- DOI: 10.1105/tpc.106.047274
Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus
Abstract
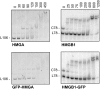
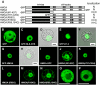
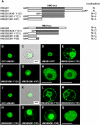
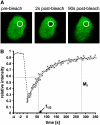
In plants, the chromatin-associated high mobility group (HMG) proteins occur in two subfamilies termed HMGA and HMGB. The HMGA proteins are characterized by the presence of four AT-hook DNA binding motifs, and the HMGB proteins contain an HMG box DNA binding domain. As architectural factors, the HMG proteins appear to be involved in the regulation of transcription and other DNA-dependent processes. We have examined the subcellular localization of Arabidopsis thaliana HMGA, HMGB1, and HMGB5, revealing that they localize to the cell nucleus. They display a speckled distribution pattern throughout the chromatin of interphase nuclei, whereas none of the proteins associate with condensed mitotic chromosomes. HMGA is targeted to the nucleus by a monopartite nuclear localization signal, while efficient nuclear accumulation of HMGB1/5 requires large portions of the basic N-terminal part of the proteins. The acidic C-terminal domain interferes with nucleolar targeting of HMGB1. Fluorescence recovery after photobleaching experiments revealed that HMGA and HMGB proteins are extremely dynamic in the nucleus, indicating that they bind chromatin only transiently before moving on to the next site, thereby continuously scanning the genome for targets. By contrast, the majority of histone H2B is basically immobile within the nucleus, while linker histone H1.2 is relatively mobile.
Figures




Similar articles
-
HMGB6 from Arabidopsis thaliana specifies a novel type of plant chromosomal HMGB protein.Biochemistry. 2004 Feb 10;43(5):1309-14. doi: 10.1021/bi035931c. Biochemistry. 2004. PMID: 14756567
-
The Arabidopsis genome encodes structurally and functionally diverse HMGB-type proteins.J Mol Biol. 2006 May 5;358(3):654-64. doi: 10.1016/j.jmb.2006.02.068. Epub 2006 Mar 10. J Mol Biol. 2006. PMID: 16563436
-
Interaction of maize chromatin-associated HMG proteins with mononucleosomes: role of core and linker histones.Biol Chem. 2003 Jul;384(7):1019-27. doi: 10.1515/BC.2003.114. Biol Chem. 2003. PMID: 12956418
-
Chromatin-associated HMGA and HMGB proteins: versatile co-regulators of DNA-dependent processes.Plant Mol Biol. 2003 Oct;53(3):281-95. doi: 10.1023/b:plan.0000007002.99408.ba. Plant Mol Biol. 2003. PMID: 14750519 Review.
-
Role of high mobility group (HMG) chromatin proteins in DNA repair.DNA Repair (Amst). 2005 Jul 28;4(8):926-38. doi: 10.1016/j.dnarep.2005.04.010. DNA Repair (Amst). 2005. PMID: 15916927 Review.
Cited by
-
High mobility group A3 enhances transcription of the DNA demethylase gene SlDML2 to promote tomato fruit ripening.Plant Physiol. 2022 May 3;189(1):315-328. doi: 10.1093/plphys/kiac063. Plant Physiol. 2022. PMID: 35171288 Free PMC article.
-
Open and closed: the roles of linker histones in plants and animals.Mol Plant. 2014 Mar;7(3):481-91. doi: 10.1093/mp/sst164. Epub 2013 Nov 22. Mol Plant. 2014. PMID: 24270504 Free PMC article.
-
Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana.Plant Mol Biol. 2009 Sep;71(1-2):39-50. doi: 10.1007/s11103-009-9507-9. Epub 2009 Jun 11. Plant Mol Biol. 2009. PMID: 19517252
-
Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains.Plant Cell. 2010 Dec;22(12):4195-215. doi: 10.1105/tpc.110.077537. Epub 2010 Dec 21. Plant Cell. 2010. PMID: 21177483 Free PMC article.
-
A Specialized Histone H1 Variant Is Required for Adaptive Responses to Complex Abiotic Stress and Related DNA Methylation in Arabidopsis.Plant Physiol. 2015 Nov;169(3):2080-101. doi: 10.1104/pp.15.00493. Epub 2015 Sep 8. Plant Physiol. 2015. PMID: 26351307 Free PMC article.
References
-
- Agresti, A., Scaffidi, P., Riva, A., Caiolfa, V.R., and Bianchi, M.E. (2005). GR and HMGB1 interact only within chromatin and influence each other's residence time. Mol. Cell 18 109–121. - PubMed
-
- Amirand, C., Viari, A., Ballini, J.-P., Rezaei, H., Beaujean, N., Jullien, D., Käs, E., and Debey, P. (1998). Three distinct sub-nuclear populations of HMG-I protein of different properties revealed by co-localization image analysis. J. Cell Sci. 111 3551–3561. - PubMed
-
- Arwood, L.J., and Spiker, S. (1990). Binding of wheat and chicken high mobility group chromosomal proteins to DNA and to wheat and chicken mononucleosomes. J. Biol. Chem. 265 9771–9777. - PubMed
-
- Bianchi, M.E., and Agresti, A. (2005). HMG proteins: Dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 15 496–506. - PubMed
-
- Boisnard-Loriq, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J., and Berger, F. (2001). Dynamic analyses of the expression of the HISTONE∷YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

