Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter
- PMID: 17113386
- PMCID: PMC1712676
- DOI: 10.1016/j.cub.2006.09.035
Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter
Abstract
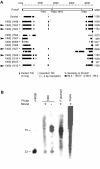
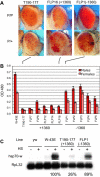
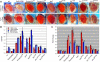
In eukaryotes, distinct regions of the genome are packaged as euchromatin (less condensed, more active) or heterochromatin (condensed, silenced). Studies in yeast, plants, and flies suggest that RNA interference (RNAi) is linked to heterochromatin formation and transcriptional silencing of transposable element (TE) sequences. We previously reported that insertion of a mobile hsp70-white reporter within 10 kb of a 1360 element on chromosome four of Drosophila melanogaster correlates with variegation (silencing). Here, we report small RNAs (approximately 23 nt) corresponding to 1360, indicating processing by the RNAi machinery. To directly test the ability of 1360 to silence a nearby gene in vivo, we introduced a P element construct carrying a single copy of 1360 upstream of the hsp70-white reporter into flies. This 1360 element contributes to HP1-dependent variegation at a pericentric insertion site, as demonstrated by a decrease in silencing after FLP-mediated removal of 1360. In euchromatin, 1360 is not sufficient to induce silencing, suggesting that proximity to pericentric heterochromatin and/or a high local TE density contributes to heterochromatin formation. Silencing of the 1360, hsp70-white reporter is sensitive to mutations in RNAi components. Our results implicate 1360 as a target for sequence-specific heterochromatic silencing through an RNAi-dependent mechanism.
Figures

Similar articles
-
Targeting of P-Element Reporters to Heterochromatic Domains by Transposable Element 1360 in Drosophila melanogaster.Genetics. 2016 Feb;202(2):565-82. doi: 10.1534/genetics.115.183228. Epub 2015 Dec 17. Genetics. 2016. PMID: 26680659 Free PMC article.
-
cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four.Mol Cell Biol. 2004 Sep;24(18):8210-20. doi: 10.1128/MCB.24.18.8210-8220.2004. Mol Cell Biol. 2004. PMID: 15340080 Free PMC article.
-
Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin.Chromosoma. 1998 Nov;107(5):277-85. doi: 10.1007/s004120050309. Chromosoma. 1998. PMID: 9880760
-
RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe.Trends Genet. 2005 Aug;21(8):450-6. doi: 10.1016/j.tig.2005.06.005. Trends Genet. 2005. PMID: 15979194 Review.
-
Analyzing heterochromatin formation using chromosome 4 of Drosophila melanogaster.Cold Spring Harb Symp Quant Biol. 2004;69:267-72. doi: 10.1101/sqb.2004.69.267. Cold Spring Harb Symp Quant Biol. 2004. PMID: 16117658 Review. No abstract available.
Cited by
-
Small RNA-directed heterochromatin formation in the context of development: what flies might learn from fission yeast.Biochim Biophys Acta. 2009 Jan;1789(1):3-16. doi: 10.1016/j.bbagrm.2008.08.002. Epub 2008 Aug 16. Biochim Biophys Acta. 2009. PMID: 18789407 Free PMC article. Review.
-
Transgenic epigenetics: using transgenic organisms to examine epigenetic phenomena.Genet Res Int. 2012;2012:689819. doi: 10.1155/2012/689819. Epub 2012 Mar 27. Genet Res Int. 2012. PMID: 22567397 Free PMC article.
-
Polycomb group-dependent, heterochromatin protein 1-independent, chromatin structures silence retrotransposons in somatic tissues outside ovaries.DNA Res. 2011 Dec;18(6):451-61. doi: 10.1093/dnares/dsr031. Epub 2011 Sep 9. DNA Res. 2011. PMID: 21908513 Free PMC article.
-
A distinct type of heterochromatin at the telomeric region of the Drosophila melanogaster Y chromosome.PLoS One. 2014 Jan 24;9(1):e86451. doi: 10.1371/journal.pone.0086451. eCollection 2014. PLoS One. 2014. PMID: 24475122 Free PMC article.
-
Chromatin domains in higher eukaryotes: insights from genome-wide mapping studies.Chromosoma. 2009 Feb;118(1):25-36. doi: 10.1007/s00412-008-0186-0. Epub 2008 Oct 14. Chromosoma. 2009. PMID: 18853173 Review.
References
-
- Haynes KA, Leibovitch BA, Rangwala SH, Craig C, Elgin SC. Analyzing heterochromatin formation using chromosome 4 of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 2004;69:267–272. - PubMed
-
- Elgin SC, Grewal SI. Heterochromatin: silence is golden. Curr Biol. 2003;13:R895–898. - PubMed
-
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. - PubMed
-
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

