Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen
- PMID: 17108083
- PMCID: PMC1838714
- DOI: 10.1073/pnas.0608595103
Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen
Abstract
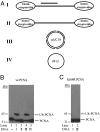
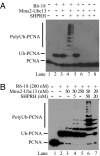
Human SHPRH gene is located at the 6q24 chromosomal region, and loss of heterozygosity in this region is seen in a wide variety of cancers. SHPRH is a member of the SWI/SNF family of ATPases/helicases, and it possesses a C(3)HC(4) RING motif characteristic of ubiquitin ligase proteins. In both of these features, SHPRH resembles the yeast Rad5 protein, which, together with Mms2-Ubc13, promotes replication through DNA lesions via an error-free postreplicational repair pathway. Genetic evidence in yeast has indicated a role for Rad5 as a ubiquitin ligase in mediating the Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Here we show that SHPRH is a functional homolog of Rad5. Similar to Rad5, SHPRH physically interacts with the Rad6-Rad18 and Mms2-Ubc13 complexes, and we show that SHPRH protein is a ubiquitin ligase indispensable for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Based on these observations, we predict a role for SHPRH in promoting error-free replication through DNA lesions. Such a role for SHPRH is consistent with the observation that this gene is mutated in a number of cancer cell lines, including those from melanomas and ovarian cancers, which raises the strong possibility that SHPRH function is an important deterrent to mutagenesis and carcinogenesis in humans.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

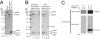
Similar articles
-
Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination.J Cell Biol. 2006 Dec 4;175(5):703-8. doi: 10.1083/jcb.200606145. Epub 2006 Nov 27. J Cell Biol. 2006. PMID: 17130289 Free PMC article.
-
Synthesis of free and proliferating cell nuclear antigen-bound polyubiquitin chains by the RING E3 ubiquitin ligase Rad5.J Biol Chem. 2009 Oct 23;284(43):29326-34. doi: 10.1074/jbc.M109.043885. Epub 2009 Aug 25. J Biol Chem. 2009. PMID: 19706603 Free PMC article.
-
Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae.Mol Cell Biol. 2006 Oct;26(20):7783-90. doi: 10.1128/MCB.01260-06. Epub 2006 Aug 14. Mol Cell Biol. 2006. PMID: 16908531 Free PMC article.
-
Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance.DNA Repair (Amst). 2010 Mar 2;9(3):257-67. doi: 10.1016/j.dnarep.2009.12.013. Epub 2010 Jan 21. DNA Repair (Amst). 2010. PMID: 20096653 Review.
-
Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA.Mutat Res. 2017 Oct;803-805:82-88. doi: 10.1016/j.mrfmmm.2017.06.004. Epub 2017 Jun 21. Mutat Res. 2017. PMID: 28666590 Review.
Cited by
-
PHD domain from human SHPRH.J Biomol NMR. 2013 Aug;56(4):393-9. doi: 10.1007/s10858-013-9758-2. Epub 2013 Aug 2. J Biomol NMR. 2013. PMID: 23907177 Free PMC article.
-
Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling.Nat Biotechnol. 2010 Aug;28(8):868-73. doi: 10.1038/nbt.1654. Epub 2010 Jul 18. Nat Biotechnol. 2010. PMID: 20639865 Free PMC article.
-
Dynamic regulation of PCNA ubiquitylation/deubiquitylation.FEBS Lett. 2011 Sep 16;585(18):2780-5. doi: 10.1016/j.febslet.2011.05.053. Epub 2011 Jun 1. FEBS Lett. 2011. PMID: 21640107 Free PMC article. Review.
-
Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks.Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12411-6. doi: 10.1073/pnas.0805685105. Epub 2008 Aug 21. Proc Natl Acad Sci U S A. 2008. PMID: 18719106 Free PMC article.
-
UBE2N Regulates Paclitaxel Sensitivity of Ovarian Cancer via Fos/P53 Axis.Onco Targets Ther. 2020 Dec 14;13:12751-12761. doi: 10.2147/OTT.S271164. eCollection 2020. Onco Targets Ther. 2020. PMID: 33363381 Free PMC article.
References
-
- Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Genes Dev. 1994;8:811–820. - PubMed
-
- Bailly V, Lauder S, Prakash S, Prakash L. J Biol Chem. 1997;272:23360–23365. - PubMed
-
- Prakash S, Johnson RE, Prakash L. Annu Rev Biochem. 2005;74:317–353. - PubMed
-
- Johnson RE, Prakash S, Prakash L. Science. 1999;283:1001–1004. - PubMed
-
- Stary A, Kannouche P, Lehmann AR, Sarasin A. J Biol Chem. 2003;278:18767–18775. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

