Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1
- PMID: 17101799
- PMCID: PMC1800796
- DOI: 10.1128/MCB.01034-06
Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1
Abstract
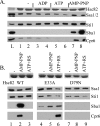
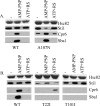
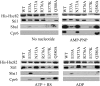
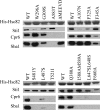
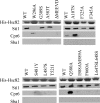
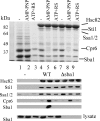
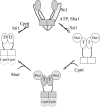
The ATP-dependent molecular chaperone Hsp90 and partner cochaperone proteins are required for the folding and activity of diverse cellular client proteins, including steroid hormone receptors and multiple oncogenic kinases. Hsp90 undergoes nucleotide-dependent conformational changes, but little is known about how these changes are coupled to client protein activation. In order to clarify how nucleotides affect Hsp90 interactions with cochaperone proteins, we monitored assembly of wild-type and mutant Hsp90 with Sti1, Sba1, and Cpr6 in Saccharomyces cerevisiae cell extracts. Wild-type Hsp90 bound Sti1 in a nucleotide-independent manner, while Sba1 and Cpr6 specifically and independently interacted with Hsp90 in the presence of the nonhydrolyzable analog of ATP, AMP-PNP. Alterations in Hsp90 residues that contribute to ATP binding or hydrolysis prevented or altered Sba1 and Cpr6 interaction; additional alterations affected the specificity of Cpr6 interaction. Some mutant forms of Hsp90 also displayed reduced Sti1 interaction in the presence of a nucleotide. These studies indicate that cycling of Hsp90 between the nucleotide-free, open conformation and the ATP-bound, closed conformation is influenced by residues both within and outside the N-terminal ATPase domain and that these conformational changes have dramatic effects on interaction with cochaperone proteins.
Figures
Similar articles
-
The Co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle.J Mol Biol. 2004 Oct 1;342(5):1403-13. doi: 10.1016/j.jmb.2004.07.064. J Mol Biol. 2004. PMID: 15364569
-
Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle.J Biol Chem. 2004 Dec 10;279(50):51989-98. doi: 10.1074/jbc.M410562200. Epub 2004 Oct 2. J Biol Chem. 2004. PMID: 15466438
-
Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones.EMBO J. 1999 Feb 1;18(3):754-62. doi: 10.1093/emboj/18.3.754. EMBO J. 1999. PMID: 9927435 Free PMC article.
-
Structure and mechanism of the Hsp90 molecular chaperone machinery.Annu Rev Biochem. 2006;75:271-94. doi: 10.1146/annurev.biochem.75.103004.142738. Annu Rev Biochem. 2006. PMID: 16756493 Review.
-
Cdc37 regulation of the kinome: when to hold 'em and when to fold 'em.Sci STKE. 2007 May 8;2007(385):pe22. doi: 10.1126/stke.3852007pe22. Sci STKE. 2007. PMID: 17488976 Review.
Cited by
-
Dynamics of the regulation of Hsp90 by the co-chaperone Sti1.EMBO J. 2012 Mar 21;31(6):1518-28. doi: 10.1038/emboj.2012.37. Epub 2012 Feb 21. EMBO J. 2012. PMID: 22354036 Free PMC article.
-
Modeling signal propagation mechanisms and ligand-based conformational dynamics of the Hsp90 molecular chaperone full-length dimer.PLoS Comput Biol. 2009 Mar;5(3):e1000323. doi: 10.1371/journal.pcbi.1000323. Epub 2009 Mar 20. PLoS Comput Biol. 2009. PMID: 19300478 Free PMC article.
-
Hsp90 mutants with distinct defects provide novel insights into cochaperone regulation of the folding cycle.PLoS Genet. 2023 May 25;19(5):e1010772. doi: 10.1371/journal.pgen.1010772. eCollection 2023 May. PLoS Genet. 2023. PMID: 37228112 Free PMC article.
-
Contributions of co-chaperones and post-translational modifications towards Hsp90 drug sensitivity.Future Med Chem. 2013 Jun;5(9):1059-71. doi: 10.4155/fmc.13.88. Future Med Chem. 2013. PMID: 23734688 Free PMC article. Review.
-
Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions.Biochem J. 2007 May 15;404(1):159-67. doi: 10.1042/BJ20070084. Biochem J. 2007. PMID: 17300223 Free PMC article.
References
-
- Chen, S., and D. F. Smith. 1998. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 273:35194-35200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
