The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes
- PMID: 17099692
- PMCID: PMC1794701
- DOI: 10.1038/sj.embor.7400828
The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes
Abstract
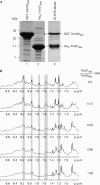
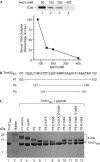
Proteins destined for the mitochondrial matrix are imported by the translocase of the outer membrane--the TOM complex--and the presequence translocase of the inner membrane--the TIM23 complex. At present, there is no structural information on components of the presequence translocase. Tim21, a subunit of the presequence translocase consisting of a membrane anchor and a carboxy-terminal domain exposed to the intermembrane space, directly connects the TOM and TIM23 complexes by binding to the intermembrane space domain of the Tom22 receptor. We crystallized the binding domain of Tim21 of Saccharomyces cerevisiae and determined its structure at 1.6 A resolution. The Tim21 structure represents a new alpha/beta-mixed protein fold with two alpha-helices flanked by an extended eight-stranded beta-sheet. We also identified a core sequence of Tom22 that binds to Tim21. Furthermore, negatively charged amino-acid residues of Tom22 are important for binding to Tim21. Here we suggest a mechanism for the TOM-TIM interaction.
Figures
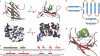
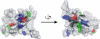
Similar articles
-
Role of Tim21 in mitochondrial translocation contact sites.J Biol Chem. 2005 Jun 24;280(25):23437-40. doi: 10.1074/jbc.C500135200. Epub 2005 May 4. J Biol Chem. 2005. PMID: 15878866
-
Molecular basis of the dynamic structure of the TIM23 complex in the mitochondrial intermembrane space.Structure. 2014 Oct 7;22(10):1501-11. doi: 10.1016/j.str.2014.07.015. Epub 2014 Sep 25. Structure. 2014. PMID: 25263020
-
Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17.Cell. 2005 Mar 25;120(6):817-29. doi: 10.1016/j.cell.2005.01.011. Cell. 2005. PMID: 15797382
-
The mitochondrial import machinery: preprotein-conducting channels with binding sites for presequences.Biochim Biophys Acta. 2002 Sep 2;1592(1):15-24. doi: 10.1016/s0167-4889(02)00260-4. Biochim Biophys Acta. 2002. PMID: 12191764 Review.
-
The preprotein translocase of the mitochondrial inner membrane: function and evolution.J Mol Biol. 1999 Feb 12;286(1):105-20. doi: 10.1006/jmbi.1998.2455. J Mol Biol. 1999. PMID: 9931253 Review.
Cited by
-
Building Better Barrels - β-barrel Biogenesis and Insertion in Bacteria and Mitochondria.J Mol Biol. 2021 Aug 6;433(16):166894. doi: 10.1016/j.jmb.2021.166894. Epub 2021 Feb 24. J Mol Biol. 2021. PMID: 33639212 Free PMC article. Review.
-
NLG1, encoding a mitochondrial membrane protein, controls leaf and grain development in rice.BMC Plant Biol. 2023 Sep 9;23(1):418. doi: 10.1186/s12870-023-04417-2. BMC Plant Biol. 2023. PMID: 37689677 Free PMC article.
-
Multiple pathways for sorting mitochondrial precursor proteins.EMBO Rep. 2008 Jan;9(1):42-9. doi: 10.1038/sj.embor.7401126. EMBO Rep. 2008. PMID: 18174896 Free PMC article. Review.
-
Mapping protein interactions in the active TOM-TIM23 supercomplex.Nat Commun. 2021 Sep 29;12(1):5715. doi: 10.1038/s41467-021-26016-1. Nat Commun. 2021. PMID: 34588454 Free PMC article.
-
Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase.Mol Biol Cell. 2008 Jun;19(6):2642-9. doi: 10.1091/mbc.e07-12-1226. Epub 2008 Apr 9. Mol Biol Cell. 2008. PMID: 18400944 Free PMC article.
References
-
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nisnikawa S-I, Endo T, Kohda D (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100: 551–560 - PubMed
-
- Brunger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 - PubMed
-
- Chacinska A et al. (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120: 817–829 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

