Predicting domain-domain interactions using a parsimony approach
- PMID: 17094802
- PMCID: PMC1794579
- DOI: 10.1186/gb-2006-7-11-r104
Predicting domain-domain interactions using a parsimony approach
Abstract
We propose a novel approach to predict domain-domain interactions from a protein-protein interaction network. In our method we apply a parsimony-driven explanation of the network, where the domain interactions are inferred using linear programming optimization, and false positives in the protein network are handled by a probabilistic construction. This method outperforms previous approaches by a considerable margin. The results indicate that the parsimony principle provides a correct approach for detecting domain-domain contacts.
Figures
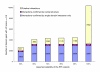
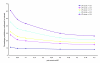
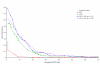
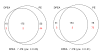
Similar articles
-
Interrogating domain-domain interactions with parsimony based approaches.BMC Bioinformatics. 2008 Mar 26;9:171. doi: 10.1186/1471-2105-9-171. BMC Bioinformatics. 2008. PMID: 18366803 Free PMC article.
-
Inferring protein domain interactions from databases of interacting proteins.Genome Biol. 2005;6(10):R89. doi: 10.1186/gb-2005-6-10-r89. Epub 2005 Sep 19. Genome Biol. 2005. PMID: 16207360 Free PMC article.
-
A discriminative approach for identifying domain-domain interactions from protein-protein interactions.Proteins. 2010 Apr;78(5):1243-53. doi: 10.1002/prot.22643. Proteins. 2010. PMID: 20027642
-
Topology and weights in a protein domain interaction network--a novel way to predict protein interactions.BMC Genomics. 2006 May 23;7:122. doi: 10.1186/1471-2164-7-122. BMC Genomics. 2006. PMID: 16716232 Free PMC article.
-
Kernel methods for predicting protein-protein interactions.Bioinformatics. 2005 Jun;21 Suppl 1:i38-46. doi: 10.1093/bioinformatics/bti1016. Bioinformatics. 2005. PMID: 15961482
Cited by
-
Translating Mendelian and complex inheritance of Alzheimer's disease genes for predicting unique personal genome variants.J Am Med Inform Assoc. 2012 Mar-Apr;19(2):306-16. doi: 10.1136/amiajnl-2011-000656. J Am Med Inform Assoc. 2012. PMID: 22319180 Free PMC article.
-
A top-down approach to infer and compare domain-domain interactions across eight model organisms.PLoS One. 2009;4(3):e5096. doi: 10.1371/journal.pone.0005096. Epub 2009 Mar 31. PLoS One. 2009. PMID: 19333396 Free PMC article.
-
DASMI: exchanging, annotating and assessing molecular interaction data.Bioinformatics. 2009 May 15;25(10):1321-8. doi: 10.1093/bioinformatics/btp142. Bioinformatics. 2009. PMID: 19420069 Free PMC article.
-
Interrogating domain-domain interactions with parsimony based approaches.BMC Bioinformatics. 2008 Mar 26;9:171. doi: 10.1186/1471-2105-9-171. BMC Bioinformatics. 2008. PMID: 18366803 Free PMC article.
-
NetGrep: fast network schema searches in interactomes.Genome Biol. 2008;9(9):R138. doi: 10.1186/gb-2008-9-9-r138. Epub 2008 Sep 18. Genome Biol. 2008. PMID: 18801179 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

