Structure and function of eukaryotic Ribonuclease P RNA
- PMID: 17081993
- PMCID: PMC1716732
- DOI: 10.1016/j.molcel.2006.09.011
Structure and function of eukaryotic Ribonuclease P RNA
Abstract
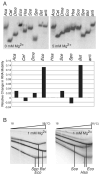
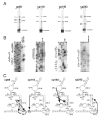
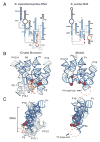
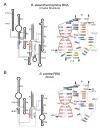
Ribonuclease P (RNase P) is the ribonucleoprotein endonuclease that processes the 5' ends of precursor tRNAs. Bacterial and eukaryal RNase P RNAs had the same primordial ancestor; however, they were molded differently by evolution. RNase P RNAs of eukaryotes, in contrast to bacterial RNAs, are not catalytically active in vitro without proteins. By comparing the bacterial and eukaryal RNAs, we can begin to understand the transitions made between the RNA and protein-dominated worlds. We report, based on crosslinking studies, that eukaryal RNAs, although catalytically inactive alone, fold into functional forms and specifically bind tRNA even in the absence of proteins. Based on the crosslinking results and crystal structures of bacterial RNAs, we develop a tertiary structure model of the eukaryal RNase P RNA. The eukaryal RNA contains a core structure similar to the bacterial RNA but lacks specific features that in bacterial RNAs contribute to catalysis and global stability of tertiary structure.
Figures

Similar articles
-
Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA.RNA. 2000 Dec;6(12):1895-904. doi: 10.1017/s1355838200001461. RNA. 2000. PMID: 11142387 Free PMC article.
-
Analysis of the tertiary structure of the ribonuclease P ribozyme-substrate complex by site-specific photoaffinity crosslinking.RNA. 1997 Jun;3(6):561-76. RNA. 1997. PMID: 9174092 Free PMC article.
-
Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs.EMBO J. 1998 Mar 2;17(5):1515-25. doi: 10.1093/emboj/17.5.1515. EMBO J. 1998. PMID: 9482748 Free PMC article.
-
RNase P: interface of the RNA and protein worlds.Trends Biochem Sci. 2006 Jun;31(6):333-41. doi: 10.1016/j.tibs.2006.04.007. Epub 2006 May 6. Trends Biochem Sci. 2006. PMID: 16679018 Review.
-
Ribonuclease P: unity and diversity in a tRNA processing ribozyme.Annu Rev Biochem. 1998;67:153-80. doi: 10.1146/annurev.biochem.67.1.153. Annu Rev Biochem. 1998. PMID: 9759486 Review.
Cited by
-
Closing the circle: replicating RNA with RNA.Cold Spring Harb Perspect Biol. 2010 Oct;2(10):a002204. doi: 10.1101/cshperspect.a002204. Epub 2010 Jun 16. Cold Spring Harb Perspect Biol. 2010. PMID: 20554706 Free PMC article. Review.
-
tRNA biology charges to the front.Genes Dev. 2010 Sep 1;24(17):1832-60. doi: 10.1101/gad.1956510. Genes Dev. 2010. PMID: 20810645 Free PMC article. Review.
-
Human RNase P: a tRNA-processing enzyme and transcription factor.Nucleic Acids Res. 2007;35(11):3519-24. doi: 10.1093/nar/gkm071. Epub 2007 May 5. Nucleic Acids Res. 2007. PMID: 17483522 Free PMC article. Review.
-
The cellular landscape of mid-size noncoding RNA.Wiley Interdiscip Rev RNA. 2019 Jul;10(4):e1530. doi: 10.1002/wrna.1530. Epub 2019 Mar 6. Wiley Interdiscip Rev RNA. 2019. PMID: 30843375 Free PMC article. Review.
-
Physical evidence supporting a ribosomal shunting mechanism of translation initiation for BACE1 mRNA.Translation (Austin). 2013 Apr 1;1(1):e24400. doi: 10.4161/trla.24400. eCollection 2013. Translation (Austin). 2013. PMID: 26824018 Free PMC article.
References
-
- Beebe JA, Kurz JC, Fierke CA. Magnesium ions are required by Bacillus subtilis ribonuclease P RNA for both binding and cleaving precursor tRNAAsp. Biochemistry. 1996;36:10493–10505. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

