Cockayne syndrome exhibits dysregulation of p21 and other gene products that may be independent of transcription-coupled repair
- PMID: 17055654
- PMCID: PMC2100027
- DOI: 10.1016/j.neuroscience.2006.08.074
Cockayne syndrome exhibits dysregulation of p21 and other gene products that may be independent of transcription-coupled repair
Abstract
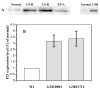
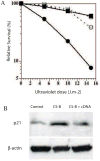
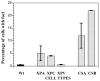
Cockayne syndrome (CS) is a progressive childhood neurodegenerative disorder associated with a DNA repair defect caused by mutations in either of two genes, CSA and CSB. These genes are involved in nucleotide excision repair (NER) of DNA damage from ultraviolet (UV) light, other bulky chemical adducts and reactive oxygen in transcriptionally active genes (transcription-coupled repair, TCR). For a long period it has been assumed that the symptoms of CS patients are all due to reduced TCR of endogenous DNA damage in the brain, together with unexplained unique sensitivity of specific neural cells in the cerebellum. Not all the symptoms of CS patients are however easily related to repair deficiencies, so we hypothesize that there are additional pathways relevant to the disease, particularly those that are downstream consequences of a common defect in the E3 ubiquitin ligase associated with the CSA and CSB gene products. We have found that the CSB defect results in altered expression of anti-angiogenic and cell cycle genes and proteins at the level of both gene expression and protein lifetime. We find an over-abundance of p21 due to reduced protein turnover, possibly due to the loss of activity of the CSA/CSB E3 ubiquitylation pathway. Increased levels of p21 can result in growth inhibition, reduced repair from the p21-PCNA interaction, and increased generation of reactive oxygen. Consistent with increased reactive oxygen levels we find that CS-A and -B cells grown under ambient oxygen show increased DNA breakage, as compared with xeroderma pigmentosum cells. Thus the complex symptoms of CS may be due to multiple, independent downstream targets of the E3 ubiquitylation system that results in increased DNA damage, reduced transcription coupled repair, and inhibition of cell cycle progression and growth.
Figures


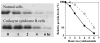
Similar articles
-
The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair.Mol Biol Cell. 1999 Nov;10(11):3583-94. doi: 10.1091/mbc.10.11.3583. Mol Biol Cell. 1999. PMID: 10564257 Free PMC article.
-
Mutations in Cockayne Syndrome-Associated Genes (Csa and Csb) Predispose to Cisplatin-Induced Hearing Loss in Mice.J Neurosci. 2016 Apr 27;36(17):4758-70. doi: 10.1523/JNEUROSCI.3890-15.2016. J Neurosci. 2016. PMID: 27122034 Free PMC article.
-
Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells.Nucleic Acids Res. 2002 Feb 1;30(3):782-93. doi: 10.1093/nar/30.3.782. Nucleic Acids Res. 2002. PMID: 11809892 Free PMC article.
-
Cockayne syndrome group B cellular and biochemical functions.Am J Hum Genet. 2003 Dec;73(6):1217-39. doi: 10.1086/380399. Epub 2003 Nov 24. Am J Hum Genet. 2003. PMID: 14639525 Free PMC article. Review.
-
Cockayne syndrome group B (CSB) protein: at the crossroads of transcriptional networks.Mech Ageing Dev. 2013 May-Jun;134(5-6):234-42. doi: 10.1016/j.mad.2013.03.004. Epub 2013 Apr 3. Mech Ageing Dev. 2013. PMID: 23562425 Review.
Cited by
-
High-confidence cancer patient stratification through multiomics investigation of DNA repair disorders.Cell Death Dis. 2022 Nov 26;13(11):999. doi: 10.1038/s41419-022-05437-w. Cell Death Dis. 2022. PMID: 36435816 Free PMC article.
-
Sirt1 suppresses RNA synthesis after UV irradiation in combined xeroderma pigmentosum group D/Cockayne syndrome (XP-D/CS) cells.Proc Natl Acad Sci U S A. 2013 Jan 15;110(3):E212-20. doi: 10.1073/pnas.1213076110. Epub 2012 Dec 24. Proc Natl Acad Sci U S A. 2013. PMID: 23267107 Free PMC article.
-
Identification and Characterization of a Novel Recurrent ERCC6 Variant in Patients with a Severe Form of Cockayne Syndrome B.Genes (Basel). 2021 Nov 29;12(12):1922. doi: 10.3390/genes12121922. Genes (Basel). 2021. PMID: 34946871 Free PMC article.
-
CSA and CSB proteins interact with p53 and regulate its Mdm2-dependent ubiquitination.Cell Cycle. 2011 Nov 1;10(21):3719-30. doi: 10.4161/cc.10.21.17905. Epub 2011 Nov 1. Cell Cycle. 2011. PMID: 22032989 Free PMC article.
-
CSB promoter downregulation via histone H3 hypoacetylation is an early determinant of replicative senescence.Nat Commun. 2019 Dec 6;10(1):5576. doi: 10.1038/s41467-019-13314-y. Nat Commun. 2019. PMID: 31811121 Free PMC article.
References
-
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. - PubMed
-
- Barrett SF, Robbins JH, Tarone RE, Kraemer KH. Evidence for defective repair of cyclobutane dimers with normal repair of other photoproducts in a transcriptionally active gene transfected into Cockayne syndrome cells. Mutation Research. 1991;255:281–291. - PubMed
-
- Beerens N, Hoeijmakers JH, Kanaar R, Vermeulen W, Wyman C. The CSB Protein Actively Wraps DNA. J Biol Chem. 2005;280:4722–4729. - PubMed
-
- Bendjennat M, Boulaire JRM, Jascur T, Brickner H, Barbier V, Sarasin A, Fotedar A, Fotedar R. UV Irradiation triggers Ubiquitin-Dependent degradation of p21 WAF1 to promote DNA repair. Cell. 2003;114:599–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

