Seven Dictyostelium discoideum phosphodiesterases degrade three pools of cAMP and cGMP
- PMID: 17040207
- PMCID: PMC1783984
- DOI: 10.1042/BJ20061153
Seven Dictyostelium discoideum phosphodiesterases degrade three pools of cAMP and cGMP
Abstract
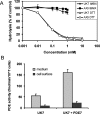
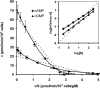
The Dictyostelium discoideum genome uncovers seven cyclic nucleotide PDEs (phosphodiesterases), of which six have been characterized previously and the seventh is characterized in the present paper. Three enzymes belong to the ubiquitous class I PDEs, common in all eukaryotes, whereas four enzymes belong to the rare class II PDEs that are present in bacteria and lower eukaryotes. Since all D. discoideum PDEs are now characterized we have calculated the contribution of each enzyme in the degradation of the three important pools of cyclic nucleotides: (i) extracellular cAMP that induces chemotaxis during aggregation and differentiation in slugs; (ii) intracellular cAMP that mediates development; and (iii) intracellular cGMP that mediates chemotaxis. It appears that each cyclic nucleotide pool is degraded by a combination of enzymes that have different affinities, allowing a broad range of substrate concentrations to be degraded with first-order kinetics. Extracellular cAMP is degraded predominantly by the class II high-affinity enzyme DdPDE1 and its close homologue DdPDE7, and in the multicellular stage also by the low-affinity transmembrane class I enzyme DdPDE4. Intracellular cAMP is degraded by the DdPDE2, a class I enzyme regulated by histidine kinase/phospho-relay, and by the cAMP-/cGMP-stimulated class II DdPDE6. Finally, basal intracellular cGMP is degraded predominantly by the high-affinity class I DdPDE3, while the elevated cGMP levels that arise after receptor stimulation are degraded predominantly by a cGMP-stimulated cGMP-specific class II DdPDE5. The analysis shows that the combination of enzymes is tuned to keep the concentration and lifetime of the substrate within a functional range.
Figures

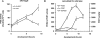

Similar articles
-
DdPDE4, a novel cAMP-specific phosphodiesterase at the surface of dictyostelium cells.J Biol Chem. 2006 Jul 21;281(29):20018-26. doi: 10.1074/jbc.M600040200. Epub 2006 Apr 27. J Biol Chem. 2006. PMID: 16644729
-
Characterization of a novel cAMP-binding, cAMP-specific cyclic nucleotide phosphodiesterase (TcrPDEB1) from Trypanosoma cruzi.Biochem J. 2006 Oct 15;399(2):305-14. doi: 10.1042/BJ20060757. Biochem J. 2006. PMID: 16776650 Free PMC article.
-
[Phosphodiesterases of cyclic GMP].Postepy Hig Med Dosw. 2001;55(5):611-27. Postepy Hig Med Dosw. 2001. PMID: 11795198 Review. Polish.
-
The Rv0805 gene from Mycobacterium tuberculosis encodes a 3',5'-cyclic nucleotide phosphodiesterase: biochemical and mutational analysis.Biochemistry. 2005 Dec 6;44(48):15695-704. doi: 10.1021/bi0512391. Biochemistry. 2005. PMID: 16313172
-
Implications of PDE4 structure on inhibitor selectivity across PDE families.Int J Impot Res. 2004 Jun;16 Suppl 1:S24-7. doi: 10.1038/sj.ijir.3901211. Int J Impot Res. 2004. PMID: 15224132 Review.
Cited by
-
Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication.Genome Res. 2011 Nov;21(11):1882-91. doi: 10.1101/gr.121137.111. Epub 2011 Jul 14. Genome Res. 2011. PMID: 21757610 Free PMC article.
-
Direct biochemical measurements of signal relay during Dictyostelium development.J Biol Chem. 2011 Nov 4;286(44):38649-38658. doi: 10.1074/jbc.M111.284182. Epub 2011 Sep 12. J Biol Chem. 2011. PMID: 21911494 Free PMC article.
-
Intramolecular activation mechanism of the Dictyostelium LRRK2 homolog Roco protein GbpC.J Biol Chem. 2008 Oct 31;283(44):30412-20. doi: 10.1074/jbc.M804265200. Epub 2008 Aug 14. J Biol Chem. 2008. PMID: 18703517 Free PMC article.
-
Multiple Roots of Fruiting Body Formation in Amoebozoa.Genome Biol Evol. 2018 Feb 1;10(2):591-606. doi: 10.1093/gbe/evy011. Genome Biol Evol. 2018. PMID: 29378020 Free PMC article.
-
Local modulation of chemoattractant concentrations by single cells: dissection using a bulk-surface computational model.Interface Focus. 2016 Oct 6;6(5):20160036. doi: 10.1098/rsfs.2016.0036. Interface Focus. 2016. PMID: 27708760 Free PMC article.
References
-
- Pitt G. S., Milona N., Borleis J., Lin K. C., Reed R. R., Devreotes P. N. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell. 1992;69:305–315. - PubMed
-
- Soderbom F., Anjard C., Iranfar N., Fuller D., Loomis W. F. An adenylyl cyclase that functions during late development of Dictyostelium. Development. 1999;126:5463–5471. - PubMed
-
- Meima M. E., Schaap P. Fingerprinting of adenylyl cyclase activities during Dictyostelium development indicates a dominant role for adenylyl cyclase B in terminal differentiation. Dev. Biol. 1999;212:182–190. - PubMed
-
- Roelofs J., Van Haastert P. J. M. Characterization of two unusual guanylyl cyclases from Dictyostelium. J. Biol. Chem. 2002;277:9167–9174. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

